Back to Journals » Cancer Management and Research » Volume 10
U94/rep of human herpesvirus 6 inhibits proliferation, invasion, and angiogenesis of glioma
Authors Gu B, Li L, Li M , Wang J, Zhang G, Yao K, Wang S
Received 20 June 2018
Accepted for publication 27 September 2018
Published 20 November 2018 Volume 2018:10 Pages 5991—6001
DOI https://doi.org/10.2147/CMAR.S177777
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Bin Gu,1 Lingyun Li,2 Meng Li,3 Jinfeng Wang,4 Guofeng Zhang,4 Kun Yao,4 Shizhi Wang5
1Department of Neurosurgery, Zhongda Hospital, School of Medicine, Southeast University, Nanjing, China; 2Department of Developmental Genetics, Nanjing Medical University, Nanjing, China; 3Department of Neurosurgery, Suqian First Hospital, Suqian, China; 4Department of Microbiology and Immunology, Nanjing Medical University, Nanjing, China; 5Key Laboratory of Environmental Medicine Engineering, Ministry of Education, School of Public Health, Southeast University, Nanjing, China
Purpose: We previously found the involvement of human herpesvirus 6 (HHV-6) infection in the pathogenesis of glioma. U94/rep, encoded by HHV-6, has been identified to play a vital role in viral gene expression and latency. Recent studies have shown its inhibition of angiogenesis and tumorigenesis in endothelial cells and prostate cancer cell line PC3, respectively. Here, we aimed to investigate the role of U94/rep in the development and progression of glioma.
Patients and methods: A total of 20 glioma tissues with positive HHV-6 infection were used for detection of U94/rep. MTT, soft agar, propidium iodide staining, wound healing, Transwell, and chick embryo chorioallantoic membrane assays were applied for evaluation of glioma cells’ proliferation, colony formation, cell cycle, migration, invasion, and angiogenesis, respectively.
Results: U94/rep transcripts could be detected in 11 out of 20 glioma tissues with positive HHV-6 infection. Furthermore, MTT and soft agar assays revealed that overexpression of U94/rep inhibited glioma cell proliferation and colony formation, which may be attributed to the cell cycle arrest at S phase induced by U94/rep. Further analysis demonstrated that U94/rep inhibited glioma cells’ migration and invasion and ex vivo angiogenesis. Reduced expression of proangiogenic factors, vascular endothelial growth factor and basic fibroblast growth factor, and type IV collagenases, MMP-2 and MMP-9, was detected in cells overexpressing U94/rep. These decreased factors may undermine glioma cell migration, invasion, and angiogenesis.
Conclusion: Our results demonstrated that U94/rep could inhibit malignant phenotypes of glioma cells, indicating that it is a potential target for therapeutic intervention.
Keywords: HHV-6, U94/rep, tumorigenesis, glioma
Introduction
Glioma is one of the most common intracranial tumors, which has a high recurrence rate and high mortality.1–3 Glioma is a highly invasive tumor and there is often no distinct boundary between the tumor and normal brain.4,5 Surgery combined with radiotherapy and chemotherapy is the main treatment; however, the recurrence rate is high and the overall 5-year survival rate of glioma remains <5%.6 Although studies have suggested that ionizing radiation, inherited mutations of highly penetrant genes, and germline polymorphisms play critical roles in the development and progression of glioma,3 the etiology of glioma remains unclear. In recent years, accumulating evidence has suggested that viral agents are involved in the pathogenesis of glioma.7 Human herpesvirus 6 (HHV-6) is a double-stranded DNA virus belonging to the human herpesvirus β subfamily, and it mainly infects the CD4+ T-cells, monocyte–macrophage cells, and glial cells.8 HHV-6 infection has been related to a number of neurological disorders, including encephalitis,9 febrile seizures in young children,10 and mesial temporal lobe epilepsy.11 We previously provided direct evidence that HHV-6 is largely found in glioma tissues,12 in line with the findings presented by Crawford et al.13,14 HHV-6 strains are divided into two variants, HHV-6A and HHV-6B, which share a high degree of homology, but recently, they have been recognized as distinct viruses rather than viral variants.15
A unique genomic characteristic of HHV-6 is the U94/rep gene, encoded by both variants with no ortholog in other human herpesviruses. U94/rep encodes a 490-amino acid protein, sharing 24% identity with adeno-associated virus type 2 (AAV-2) Rep78/68 gene products at the amino acid level.16 AAV-2 Rep78/68 is a non-structural protein essential for AAV integration within chromosomes.17 Structural homology of U94/rep to Rep 78/68 suggests functional similarities between these proteins. The hypothesis is confirmed by the observation that U94/rep complemented the replication of rep-deficient AAV-2 genome16 and U94 inhibited gene transcription as did Rep 78/68.18 U94/rep mRNA and protein were expressed at a low level during infection, and ectopic expression of U94/rep inhibited HHV-6 lytic replication19 and in vitro angiogenesis and lymphangiogenesis.20
Previous studies demonstrated that NIH 3T3 cell line stably expressing U94 gene suppressed transformation by the oncogene H-ras18 and expression of U94 protein inhibited tumorigenesis of prostate cancer PC3 cell line.21 In the present study, we aimed to investigate the antitumor potential of U94/rep using glioma cell lines U251 and U87 stably transfected with U94/rep. Our results showed that U94/rep could inhibit the proliferation, migration, invasion, and angiogenesis of glioma cells. Vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF) are two classical proangiogenic factors which induce angiogenic signal transduction by binding to their receptors in the cells.22 MMP-2 and MMP-9, type IV collagenases, are collectively capable of degrading extracellular matrix components which have been reported to be related to cancer cell invasion, metastasis, and angiogenesis.23 We further evaluated the expression of VEGF, bFGF, MMP-2, and MMP-9 and found reduced expression of VEGF, bFGF, MMP-2, and MMP-9 in the cells overexpressing U94/rep.
Patients and methods
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board (IRB) of Southeast University and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. The study was approved by the IRB of Southeast University.
Patients and samples
A total of 20 glioma tissues with positive HHV-6 infection were used for U94/rep transcript detection. These specimens were obtained from patients undergoing surgery in the First Affiliated Hospital of Nanjing Medical University from 2008 to 2012. Each subject has signed an informed consent. The HHV-6 infection status of glioma tissues was determined using nested PCR and immunohistochemical assay which has been described elsewhere.12
Cell lines and reagents
Human glioma cell lines U251 and U87, human embryonic kidney 293T, and Escherichia coli DH5α cells were obtained from Prof Yao (Department of Microbiology and Immunology, Nanjing Medical University). The use of the cell lines was approved by the IRB of Southeast University. U251, U87, and 293 T cells were cultured in DMEM plus 10% FBS and grown at 37°C and 5% CO2.
Lentivirus vector construction
pSR2PH-U94/rep-overexpressing U94/rep was a kind gift from Prof Di Luca.19 The full-length U94/rep gene was obtained by PCR amplification and subsequently subjected to excise by XbaI and BstBI digestion and finally subcloned into the lentivirus vector p3D-EGH which was constructed by Shanghai 3D Biopharm Co., Ltd (Shanghai, China). Sequences of primers were: P1: 5′-CTAGTCTAGAXbaITTTTCCATAATAAATCCGAGTGATGATTTCTGG-3′ and P2: 5′-GCCTTCGAABstBI GTGGTGATGGTGATGATGCCGC-3′. The p3D-EGH and the latest version of HIV-based packaging system, Gag/Pol, Rev, and VSV-G were co-transfected into 293 T cells for lentivirus production.
Cells stably transfected with U94/rep lentivirus
The human glioma cell lines U251 and U87 were infected with U94/rep lentivirus vectors (referred as U251 or U87-U94/rep-GFP) and mock lentivirus vectors (referred as U251 or U87-GFP). Non-transfected cells were included as mock control. Stable cell lines expressing U94/rep were selected for 8 days with 800 µg/mL hygromycin 48 hours after infection. After 8 days of selection, the cell lysates were collected for the detection of U94/rep protein level by Western blot analysis.
MTT assay
The cells were seeded in 96-well plates and cultured for 24, 48, 72, and 96 hours. Cell culture was subsequently incubated with MTT reagent (5 mg/mL) at 37°C for 4 hours, and the absorbance at 550 nm was measured using a spectrophotometric plate reader.
Soft agar assay
A total of 5×104 cells were suspended in 0.3% agar and supplemented with DMEM containing 10% FBS and poured onto 1 mL of an 0.8% agar medium base layer in six-well plates. After 2–3 weeks, the number of colonies (>1 mm) was counted using a microscope.
Cell cycle and apoptosis analysis
The cells were washed twice with PBS and then fixed with 70% ethanol overnight at 4°C. Fixed cells were then resuspended in PBS containing propidium iodide (PI)/RNase, incubated at 4°C for 30 minutes, and finally analyzed by flow cytometry.
Wound healing assay
A total of 3×105 cells were cultured in six-well plates to form a monolayer. Cells were starved in serum-free medium for 12 hours to inactivate cell proliferation and then wounded by pipette tips. The wound was allowed to heal for 2 days in a fresh medium containing 1% FBS. Images of cells were photographed at the indicated day using fluorescent microscopy (IX70; Olympus Corporation, Tokyo, Japan) with a 10× objective. The wound healing abilities were evaluated using the index of cure rate which was defined as (T−Tt)/T×100%, where T was defined as the width of original wound, while Tt was the width of wound at the given time points.
Transwell assay
The invasion ability of cells was analyzed using 12-well Transwell chambers (Corning Incorporated, Corning, NY, USA) with a pore size of 8 µm. The lower compartment contained 0.6 mL DMEM with 10% FBS. Cells (1×105) were seeded into the upper compartment of invasion chambers. The bottom chamber was filled with 0.6 mL DMEM containing 10% FBS. After 24 hours of incubation at 37°C, the migrated cells were fixed with methanol, stained with crystal violet (0.04% in water), and counted.
Chick embryo chorioallantoic membrane (CAM) assay
Fertilized leghorn chicken eggs were candled on embryonic day 5 and a small window was made at the top of the live eggs. Cell suspensions (5×105) were mixed with Matrigel and placed on top of the CAM. The cells were from the same passages to ensure consistent cell viability among the different groups. The eggs were resealed and returned to the incubator until day 10 of chick embryo development. The eggs were then photographed by stereomicroscope, and vessels radially converging toward the center were pictured using a microscope. Three random fields around the tumors were then selected, and the primary blood vessels converging to tumors were counted.
Reverse transcription-PCR
Total RNA was isolated from glioma tissues using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription was performed using the ReverTra Ace qPCR RT Kit (Toyobo Co., Ltd, Osaka, Japan). The cDNA was amplified using PCR. The following primers were used for amplification: GCATACGTGCACCAATCATC and ACGCTAAAGCGGA GAATAAA.
Western blotting
Cells were lysed using the mammalian protein extraction reagent RIPA (Beyotime, Haimen, China). Western blots were performed as previously reported24 using antibodies of MMP-2, MMP-9, VEGF, and bFGF (Santa Cruz Biotechnology Inc., Dallas, TX, USA).
Statistical analyses
Data were presented as mean±SD and the differences were analyzed using ANOVA with Tukey’s honestly significant difference post hoc test. All statistical analyses were performed using R 3.2.5 software, and differences were considered statistically significant at P<0.05.
Results
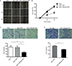
U94/rep mRNA can be detected in glioma tissues with positive HHV-6 infection
To determine whether HHV-6 is expressing U94/rep during glioma development, we tested the mRNA expression of U94/rep in the 20 glioma tissues with positive HHV-6 infection (Figure 1). The results showed that U94/rep mRNA could be detected in eleven tissues with a positive rate of 55%.
U94/rep inhibited cell proliferation and colony formation of glioma cells
U251-U94/rep-GFP cells had the lowest cell proliferation rate 3 days after transfection compared with U251 cells (mock control) and U251-GFP cells (negative control), as demonstrated by cell number counting and MTT assays (Figure 2A, B, respectively). The number of cells after 3 days of culture was (5.78±0.37)×105, (5.87±0.39)×105, and (3.77±0.44)×105 for the groups of U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in cell numbers at 72 and 96 hours after transfection between U251-U94/rep-GFP and U251 cells were statistically significant (P=0.007 and 0.003 for 72 and 96 hours, respectively). The differences in cell numbers at 72 and 96 hours after transfection between U251-U94/rep-GFP and U251-GFP cells were also statistically significant (P=0.003 and 0.001 for 72 and 96 hours, respectively). Repeating the MTT assay in another glioma U87 cell line revealed similar tendency (Figure 2C).
In addition, U251-U94/rep-GFP cells exhibited the lowest colony numbers compared with the two control cells (Figure 2D, E). The number of colonies after 2 weeks of culture was 138±12, 132±12, and 62±9 for the groups of U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in colony numbers among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=4×10−4; U251-U94/rep-GFP vs U251-GFP cells, P=6×10−4).
U94/rep induced cell cycle arrest at S phase and apoptosis in U251 cells
To determine whether U94/rep inhibited cell proliferation through cell cycle arrest, the cell cycle distribution of the three kinds of cells was detected by fluorescence-activated cell sorting after 48 hours of culture. The percentage of U251-U94/rep-GFP cells at S phase of cell cycle was significantly increased compared with the control cells. Meanwhile, a markedly decreased percentage of U251-U94/rep-GFP cells at G1 phase was observed, implying an S phase cell cycle arrest induced by U94/rep (Figure 3A–D). The percentages of G0/G1, S, and G2/M phases were 63.32%±3.83%, 27.21%±3.02%, and 9.46%±1.28%, respectively, for U251 cells; 63.68%±4.20%, 25.78%±3.49%, and 10.55%±1.63%, respectively, for U251-GFP cells; and 34.25%±2.58%, 55.10%±2.55%, and 10.65%±0.96%, respectively, for U251-U94/rep-GFP cells. The differences in G1 and S phases of cell cycle between U251-U94/rep-GFP and U251 cells were statistically significant (P=2×10−4 and 7×10−5, respectively). Similarly, the differences between U251-U94/rep-GFP and U251-GFP cells were also statistically significant (P=1×10−4 and 6×10−5, respectively). However, the differences in G2/M phase of cell cycle among the three types of cells were not statistically significant (U251-U94/rep-GFP vs U251 cells, P=0.545; U251-U94/rep-GFP vs U251-GFP cells, P=0.995).
We also evaluated glioma cell apoptosis induced by overexpression of U94/rep by PI staining. The percentage of apoptotic cells was the greatest for U251-U94/rep-GFP cells among the three types of cells (Figure 3D, the left-most column). The percentages of apoptosis cells were 0.79%±0.43%, 0.96%±0.51%, and 6.32%±1.05% for U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in apoptotic cells among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=2×10−4; U251-U94/rep-GFP vs U251-GFP cells, P=2×10−4).
U94/rep inhibited the migration and invasion of glioma cells
We also evaluated the effect of U94/rep on the migration and invasion of U251 cells. Wound healing assay showed dampened migration of U251-U94/rep-GFP cells toward the scraped clear area compared with the control cells (Figure 4A, B). The cure rate of cells at 24 hours after transfection was 34.33%±4.37%, 37.84%±3.46%, and 17.81%±3.96% for the groups of U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in cure rate among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=0.005; U251-U94/rep-GFP vs U251-GFP cells, P=0.002). The cure rate of cells at 48 hours after transfection was 62.32%±2.68%, 67.58%±3.55%, and 29.61%±3.28% for the groups of U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in cure rate among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=4×10−5; U251-U94/rep-GFP vs U251-GFP cells, P=2×10−5).
Transwell invasion assay revealed the lowest number of invasion cells in relation to U251-U94/rep-GFP cells compared with the control cells (Figure 4C). The invasion cells were 36±6, 35±5, and 17±4 for the groups of U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in invasion cells among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=0.001; U251-U94/rep-GFP vs U251-GFP cells, P=0.002). Repeating the Transwell assay in U87 cells revealed similar tendency (Figure 4D).
U94/rep suppressed angiogenesis of U251 cells
Since U94/rep was able to inhibit the proliferation, migration, and invasion of U251 cells, we further set out to determine whether U94/rep could influence angiogenesis of U251 cells. CAM assay displayed the lowest vascular density for U251-U94/rep-GFP cells than the control cells (Figure 5A, top row, B). The number of blood vessels after 10 days of culture was 8.50±1.98, 8.83±1.90, and 3.75±1.66 for the groups of U251, U251-GFP, and U251-U94/rep-GFP cells, respectively. The differences in blood vessels among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=1×10−6; U251-U94/rep-GFP vs U251-GFP cells, P=3×10−7). Furthermore, the tumor taken out from CAM infected with U251-U94/rep-GFP had the lightest red color than its counterparts (Figure 5A, bottom row), which also implied the strongest inhibition of angiogenesis. The results indicated U94/rep could effectively inhibit angiogenesis ex vivo.
U94/rep downregulated the expression of proangiogenic factors and MMPs
We analyzed the association of U94/rep with MMPs expression. In particular, the expression of MMP-2 and MMP-9 was significantly reduced in U251-U94/rep-GFP cells compared with the two control cells (Figure 6A). The differences in band intensity scores of MMP-2 and MMP-9 among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=0.002 for MMP-2 and 0.003 for MMP-9; U251-U94/rep-GFP vs U251-GFP cells, P=0.002 for MMP-2 and 0.004 for MMP-9). Because angiogenesis requires the binding of proangiogenic factors, such as VEGF and bFGF, to receptors on the surface of endothelial cells (ECs), we further evaluated the effect of U94/rep on the expression of VEGF and bFGF. As shown in Figure 6B, U251-U94/rep-GFP had the lowest expression of both VEGF and bFGF compared with the two control cells. The differences in band intensity scores of VEGF and bFGF among the three types of cells were statistically significant (U251-U94/rep-GFP vs U251 cells, P=5×10−4 for VEGF and 5×10−4 for bFGF; U251-U94/rep-GFP vs U251-GFP cells, P=7×10−4 for VEGF and 0.0005 for bFGF).
Discussion
Several studies have demonstrated that HHV-6 can be detected in primary and recurrent central nervous system tumors and more frequently in glioma. We previously provided direct evidence of existence of HHV-6 in glioma tissues and in fluid specimens from glioma cysts.12 U94/rep is a unique gene encoded by HHV-6 and is absent in all other herpesviruses. Previous research showed that NIH3T3 cells stably expressing U94 gene suppressed transformation by H-ras, indicating a tumor suppressor role of U94/rep.18 Ifon et al21 determined the antitumor potential of U94/rep in prostate cancer cell line PC3 and found U94/rep inhibited focus formation in culture and tumorigenesis in nude mice. Similarly, our study also found the antitumor activity of U94/rep which could inhibit the proliferation of glioma cells. We also determined the effect of U94/rep on the cell cycle of U251 and found that U94/rep could induce S phase cell cycle arrest in U251, which might be attributed to the antiproliferative activity of U94/rep. We previously revealed that HHV-6A induced cell apoptosis in primary human fetal astrocytes.25 Therefore, U94/rep may trigger cell apoptosis through S phase cell cycle arrest in U251 cells. Accordingly, we observed that the U251-U94/rep-GFP cells had the greatest percentage of apoptotic cells compared with the other two control cells. The extrinsic and intrinsic apoptosis pathways involving U94/rep would merit further investigation.
It is counterintuitive that HHV-6 plays an important role in glioma development, but its encoded protein U94/rep inhibited glioma growth in the present study. However, Rotola et al26 reported that the U94/rep gene product regulates viral gene expression and enables the establishment and/or maintenance of latent infection in lymphoid cells. Caselli et al19 found that U94/rep can bind ssDNA, inhibit viral DNA replication, and have a role in viral latency. In agreement with their findings, our previous study27 also found that with increase in the passage of U251 cells infected with HHV-6A, the copy number of viral DNA decreased gradually to a stable low level, whereas U94/rep, a representative of the latent stage of infection of the virus, decreased gradually and was finally maintained at a stable level. Therefore, the effect of HHV-6 infection of host cells is dependent on HHV-6 infection modes.The acute phase of HHV-6 infection could promote glioma cell proliferation and angiogenesis, whereas the latent phase of HHV-6 infection suppressed not only its own replication, but also glioma cell proliferation and angiogenesis, which may be attributed to U94/rep.
Angiogenesis is essential for tumor growth and metastasis because it can supply the necessary oxygen and nutrients to and remove wastes from tumor cells.22,28 Caruso et al20 observed that HHV-6 infection induced the loss of angiogenic properties both in lymphatic ECs and in vascular ECs, and the antiangiogenic effects were associated with U94/rep expression. In agreement with Caruso et al, we also found U94/rep could inhibit ex vivo angiogenesis in U251 cells by CAM assay. Proangiogenic factors such as VEGF and bFGF induce angiogenic signal transduction by binding to their receptors in the cells.22 We also evaluated the expression of VEGF and bFGF and found they were downregulated in the U251 cells stably expressing U94/rep compared with the control cells. VEGFs and FGFs can specifically bind to the cell surface-expressed receptors equipped with receptor kinase activity, which in turn activate downstream signal transduction pathways that regulate the proliferation, migration, and differentiation of ECs.29 The known activated intracellular signaling cascades include Ras pathway,30 Src family tyrosine kinases,31,32 phosphoinositide 3-kinase (PI3K),33,34 and the PLC pathway.33,35 Thus, the signal cascades underlying the inhibitory effect of U94/rep on glioma cells’ angiogenesis via targeting FGFs and VEGFs need further investigation. Current antiangiogenic therapies use drugs specifically targeting a single proangiogenic molecule. However, tumors, especially glioma, can activate alternative pathways to stimulate the angiogenic process, which is observed in mice wherein inhibition of VEGF can be compensated by an enhanced bFGF pathway.36 The ability of U94/rep to block both vascular and lymphatic angiogenesis opens the way to a promising field of developing drugs to prevent recurrence and metastasis of glioma which is resistant to radio- and chemotherapy.20
Invasion and metastasis are two mechanisms for cancers spreading throughout the body. In particular, MMP-2 and MMP-9 play fundamental roles in invasion and angiogenesis, and overexpression of MMP-2 and MMP-9 has been found in multiple tumors facilitating tumor cells’ migration and invasion.23 The present study displayed the lowest expression of MMP-2 and MMP-9 in the U251-U94/rep cells compared with the two control cells. A lot of classical signaling pathways have been identified to be involved in MMP9-mediated cell migration, such as JNK,37 MAPK, and PI3K signaling pathways.38 Redondo-Muñoz et al39 reported that proMMP-9 can bind to its docking receptors α4β1 integrin and CD44 and induce an intracellular signaling pathway, that is, Lyn/p-STAT3/MCL-1, which favors the survival of chronic lymphocytic leukemia cells. Another research reported that heterodimerization of proMMP-9 with CD44 can activate EGFR, which leads to phosphorylation of its downstream effectors ERK, AKT, and FAK and favors cell migration and invasion.40 Chetty et al41 found the interaction of proMMP-2 with αvβ3 induces PI3K/AKT-mediated VEGF expression and related angiogenesis. Previous studies raise the possibility that U94/rep may activate the classical signaling pathways, such as PI3K/AKT, MAPK, and EGFR pathways, involved in major biological events (cell growth, migration, invasion, and survival) to directly influence cell behavior.
The antiangiogenic properties of U94/rep are associated with the carboxy- and amino-terminal portions of U94/rep structural protein. Caruso et al20 found that transfection of plasmids deleted in the N- and C-terminal portions of U94/rep failed to inhibit angiogenesis. They also identified the nuclear localization of the U94/rep protein, and raised the possibility that U94/rep might act indirectly, inducing the expression of specific cell factors.20 The hypothesis is supported by the observation that U94/rep can bind both to single-stranded DNA42 and to transcription factor TATA-binding protein.43 The potential cell factors regulated by U94/rep remain to be identified. In addition, other functional proteins of HHV-6 might crosstalk with U94/rep and participate in the development of glioma. For example, HHV-6 DR7 (ORF1) could bind to p53 and influence cell proliferation.44,45 In our unpublished study, we also observed decreased expression of p53 and enhanced cell proliferation, invasion, and angiogenesis in the glioma cells overexpressing DR7. The interaction between U94/rep and other proteins of HHV-6 virus including DR7 warrants further investigation.
Some limitations should be addressed. First, it is a deficiency of our study that the expression of cycle-related markers such as cyclin D/E was not detected. In the present study, we mainly focus on the phenotype of glioma cell U251 overexpressing U94/rep and did not explore extensively the underlying mechanisms. We will determine the mechanisms underlying the effect of deregulated cell cycle induced by overexpressing U94/rep on glioma cell proliferation in future. Second, we simply evaluated glioma cell apoptosis induced by overexpression of U94/rep by PI staining. The results provided clues for our further investigation of extrinsic and intrinsic apoptosis pathways involving U94/rep.
Conclusion
Our data revealed that U94/rep was capable of suppressing proliferation, migration, invasion, and angiogenesis of U251 cells, suggesting U94/rep may play a tumor suppressor role in the pathogenesis of glioma as well as a potential target for therapeutic intervention.
Acknowledgment
This study was partly supported by National Natural Science Foundation of China (81301698 and 81201520), Natural Science Foundation of Jiangsu Province (BK20171367 and BK20171489), Key Project supported by Medical Science and Technology Development Foundation, Jiangsu Provincial Commission of Health and Family Planning (Z201409), and the Fundamental Research Funds for the Central Universities (2242018K40020).
Disclosure
The authors report no conflicts of interest in this work.
References
Ricard D, Idbaih A, Ducray F, Lahutte M, Hoang-Xuan K, Delattre JY. Primary brain tumours in adults. Lancet. 2012;379(9830):1984–1996. | ||
Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12(9):495–508. | ||
Schwartzbaum JA, Fisher JL, Aldape KD, Wrensch M. Epidemiology and molecular pathology of glioma. Nat Clin Pract Neurol. 2006;2(9):494–503; quiz 1 p following 516. | ||
Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–446. | ||
Diamandis P, Aldape KD. Insights from molecular profiling of adult glioma. J Clin Oncol. 2017;35(21):2386–2393. | ||
Chen J, McKay RM, Parada LF. Malignant glioma: lessons from genomics, mouse models, and stem cells. Cell. 2012;149(1):36–47. | ||
Miller G. Brain cancer. A viral link to glioblastoma? Science. 2009;323(5910):30–31. | ||
Lusso P. Human herpesvirus 6 (HHV-6). Antiviral Res. 1996;31(1–2):1–21. | ||
Rantala H, Mannonen L, Ahtiluoto S, et al. Human herpesvirus-6 associated encephalitis with subsequent infantile spasms and cerebellar astrocytoma. Dev Med Child Neurol. 2000;42(6):418–421. | ||
Cohen JI. Human herpesvirus types 6 and 7 (exanthema subitum). In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 8th ed. Vol. 2. Philadelphia: Elsevier Saunders; 2015:1772–1776. | ||
Fotheringham J, Donati D, Akhyani N, et al. Association of human herpesvirus-6B with mesial temporal lobe epilepsy. PLoS Med. 2007;4(5):e180. | ||
Chi J, Gu B, Zhang C, et al. Human herpesvirus 6 latent infection in patients with glioma. J Infect Dis. 2012;206(9):1394–1398. | ||
Crawford JR, Santi MR, Cornelison R, Sallinen SL, Haapasalo H, MacDonald TJ. Detection of human herpesvirus-6 in adult central nervous system tumors: predominance of early and late viral antigens in glial tumors. J Neurooncol. 2009;95(1):49–60. | ||
Crawford JR, Santi MR, Thorarinsdottir HK, et al. Detection of human herpesvirus-6 variants in pediatric brain tumors: association of viral antigen in low grade gliomas. J Clin Virol. 2009;46(1):37–42. | ||
Ablashi D, Agut H, Alvarez-Lafuente R, et al. Classification of HHV-6A and HHV-6B as distinct viruses. Arch Virol. 2014;159(5):863–870. | ||
Thomson BJ, Weindler FW, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204(1):304–311. | ||
Linden RM, Ward P, Giraud C, Winocour E, Berns KI. Site-specific integration by adeno-associated virus. Proc Natl Acad Sci U S A. 1996;93(21):11288–11294. | ||
Araujo JC, Doniger J, Stöppler H, Sadaie MR, Rosenthal LJ. Cell lines containing and expressing the human herpesvirus 6A ts gene are protected from both H-ras and BPV-1 transformation. Oncogene. 1997;14(8):937–943. | ||
Caselli E, Bracci A, Galvan M, et al. Human herpesvirus 6 (HHV-6) U94/REP protein inhibits betaherpesvirus replication. Virology. 2006;346(2):402–414. | ||
Caruso A, Caselli E, Fiorentini S, et al. U94 of human herpesvirus 6 inhibits in vitro angiogenesis and lymphangiogenesis. Proc Natl Acad Sci U S A. 2009;106(48):20446–20451. | ||
Ifon ET, Pang AL, Johnson W, et al. U94 alters FN1 and ANGPTL4 gene expression and inhibits tumorigenesis of prostate cancer cell line PC3. Cancer Cell Int. 2005;5:19. | ||
Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473(7347):298–307. | ||
Ala-aho R, Kähäri VM. Collagenases in cancer. Biochimie. 2005;87(3–4):273–286. | ||
Wang S, Ma G, Zhu H, et al. miR-107 regulates tumor progression by targeting NF1 in gastric cancer. Sci Rep. 2016;6:36531. | ||
Gu B, Zhang GF, Li LY, et al. Human herpesvirus 6A induces apoptosis of primary human fetal astrocytes via both caspase-dependent and -independent pathways. Virol J. 2011;8:530. | ||
Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc Natl Acad Sci U S A. 1998;95(23):13911–13916. | ||
Gu B, Zhang G, Li L, Zhou F, Feng D, Yao K. [Human herpesvirus 6A subtype latent infection of human glioma cell line U251]. Acta Univ Med Nanjing. 2015;10:1339–1343. Chinese. | ||
Albini A, Tosetti F, Li VW, Noonan DM, Li WW. Cancer prevention by targeting angiogenesis. Nat Rev Clin Oncol. 2012;9(9):498–509. | ||
Cross MJ, Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol Sci. 2001;22(4):201–207. | ||
Henkemeyer M, Rossi DJ, Holmyard DP, et al. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377(6551):695–701. | ||
Klint P, Kanda S, Kloog Y, Claesson-Welsh L. Contribution of Src and Ras pathways in FGF-2 induced endothelial cell differentiation. Oncogene. 1999;18(22):3354–3364. | ||
He H, Venema VJ, Gu X, Venema RC, Marrero MB, Caldwell RB. Vascular endothelial growth factor signals endothelial cell production of nitric oxide and prostacyclin through flk-1/KDR activation of c-Src. J Biol Chem. 1999;274(35):25130–25135. | ||
Cross MJ, Hodgkin MN, Roberts S, Landgren E, Wakelam MJ, Claesson-Welsh L. Tyrosine 766 in the fibroblast growth factor receptor-1 is required for FGF-stimulation of phospholipase C, phospholipase D, phospholipase A(2), phosphoinositide 3-kinase and cytoskeletal reorganisation in porcine aortic endothelial cells. J Cell Sci. 2000;113 (Pt 4):643–651. | ||
Gerber HP, McMurtrey A, Kowalski J, et al. Vascular endothelial growth factor regulates endothelial cell survival through the phosphatidylinositol 3’-kinase/Akt signal transduction pathway. Requirement for Flk-1/KDR activation. J Biol Chem. 1998;273(46):30336–30343. | ||
Takahashi T, Shibuya M. The 230 kDa mature form of KDR/Flk-1 (VEGF receptor-2) activates the PLC-gamma pathway and partially induces mitotic signals in NIH3T3 fibroblasts. Oncogene. 1997;14(17):2079–2089. | ||
Dempke WC, Heinemann V. Resistance to EGF-R (erbB-1) and VEGF-R modulating agents. Eur J Cancer. 2009;45(7):1117–1128. | ||
Hu X, Paik PK, Chen J, et al. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity. 2006;24(5):563–574. | ||
Dufour A, Sampson NS, Zucker S, Cao J. Role of the hemopexin domain of matrix metalloproteinases in cell migration. J Cell Physiol. 2008;217(3):643–651. | ||
Redondo-Muñoz J, Ugarte-Berzal E, Terol MJ, et al. Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia b cell survival through its hemopexin domain. Cancer Cell. 2010;17(2):160–172. | ||
Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7(9):493–507. | ||
Chetty C, Lakka SS, Bhoopathi P, Rao JS. MMP-2 alters VEGF expression via alphaVbeta3 integrin-mediated PI3K/AKT signaling in A549 lung cancer cells. Int J Cancer. 2010;127(5):1081–1095. | ||
Yoshikawa T, Asano Y, Akimoto S, et al. Latent infection of human herpesvirus 6 in astrocytoma cell line and alteration of cytokine synthesis. J Med Virol. 2002;66(4):497–505. | ||
Mori Y, Dhepakson P, Shimamoto T, et al. Expression of human herpesvirus 6B rep within infected cells and binding of its gene product to the TATA-binding protein in vitro and in vivo. J Virol. 2000;74(13):6096–6104. | ||
Kashanchi F, Araujo J, Doniger J, et al. Human herpesvirus 6 (HHV-6) ORF-1 transactivating gene exhibits malignant transforming activity and its protein binds to p53. Oncogene. 1997;14(3):359–367. | ||
Lacroix A, Collot-Teixeira S, Mardivirin L, et al. Involvement of human herpesvirus-6 variant B in classic Hodgkin’s lymphoma via DR7 oncoprotein. Clin Cancer Res. 2010;16(19):4711–4721. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.