Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 12
Type 2 Diabetes-Associated Genetic Polymorphisms as Potential Disease Predictors
Authors Witka BZ, Oktaviani DJ, Marcellino M, Barliana MI, Abdulah R
Received 6 September 2019
Accepted for publication 19 November 2019
Published 18 December 2019 Volume 2019:12 Pages 2689—2706
DOI https://doi.org/10.2147/DMSO.S230061
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Antonio Brunetti
Beska Z Witka,1 Dede J Oktaviani,1 Marcellino Marcellino,1 Melisa I Barliana,2,3 Rizky Abdulah1,3
1Departement of Pharmacology and Clinical Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Jatinangor, Indonesia; 2Departement of Biological Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Jatinangor, Indonesia; 3Center of Excellence in Higher Education for Pharmaceutical Care Innovation, Universitas Padjadjaran, Jatinangor, Indonesia
Correspondence: Melisa I Barliana
Department of Biological Pharmacy, Faculty of Pharmacy, Universitas Padjadjaran, Jl. Raya Bandung Sumedang KM. 21, Jatinangor 45363, Indonesia
Email [email protected]
Abstract: Diabetes is a major cause of mortality worldwide. There are several types of diabetes, with type 2 diabetes mellitus (T2DM) being the most common. Many factors, including environmental and genetic factors, are involved in the etiology of the disease. Numerous studies have reported the role of genetic polymorphisms in the initiation and development of T2DM. While genome-wide association studies have identified around more than 200 susceptibility loci, it remains unclear whether these loci are correlated with the pathophysiology of the disease. The present review aimed to elucidate the potential genetic mechanisms underlying T2DM. We found that some genetic polymorphisms were related to T2DM, either in the form of single-nucleotide polymorphisms or direct amino acid changes in proteins. These polymorphisms are potential predictors for the management of T2DM.
Keywords: type 2 diabetes, genetic polymorphisms, susceptibility prediction
Introduction
Diabetes is a chronic disease that can lead to serious complications. It is classified into two main types: type 1 diabetes mellitus and type 2 diabetes mellitus (T2DM). T2DM is a metabolic disorder that is characterized by peripheral insulin resistance and impaired insulin secretion.1 During the period from 1980 to 2008, the number of people worldwide with T2DM has more than doubled.2 Studies on the prevalence of diabetes in the adult population aged 20–79 years estimated that the worldwide prevalence of people with T2DM was 6.4% in 2010, where 285 million adults had T2DM. By 2030, 439 million adults are predicted to have T2DM, accounting for 7.7% of the adult population worldwide.3
Environmental and genetic factors are involved in the pathogenesis of T2DM.4 The majority of genes involved play a role in β-cell function. Genetic polymorphisms that have impacts on important proteins that participate in glucose metabolism and insulin secretion may also affect susceptibility to T2DM.5 Genome-wide association studies (GWASs), the candidate gene approach, and linkage analysis have identified various genes that contribute to T2DM susceptibility.6–8 The development of genetic risk scores using combined analysis of loci has significantly contributed to predicting the incidence of T2DM.9–11 Therefore, it is possible to facilitate early diagnosis and determine preventative strategies to reduce the incidence of the disease.12–15
T2DM has a strong genetic basis, and individuals with a first-degree family history are at increased risk of developing the disease, and this risk is increased twofold if both parents have diabetes.16 Several risk factors for T2DM have been identified, including obesity and central obesity, ethnicity, family history of diabetes, elevated blood pressure, dyslipidemia, lifestyle factors and dietary intake.17–19 Some of these risk factors are associated with functional metabolism; therefore, genetic-based diagnoses may provide a more promising diagnostic tool. More than 200 genetic loci have been detected to be associated with T2DM risk,6 the genes elaborated in this review represent only a selected subset of T2DM-associated genes.
Methodology
The present review included studies published in the PubMed database obtained using the keywords “gene prediction”, “gene association”, and “type 2 diabetes”. Reviews, non-English studies, unrelated studies, such as non-human studies and reporting T2DM complications, were excluded. A flowchart of the literature search is shown in Figure 1.
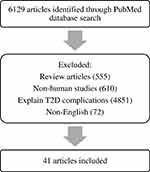 |
Figure 1 Flowchart outlining the literature search process. |
Of the 6129 articles obtained in June 2019, we included 41 studies that focused specifically on the association between genetics and the prediction of T2DM (Table 1), where several genes have been associated with T2DM and can be used as predictors of the disease, including KLF14, KCNQ1, DUSP9, FTO, HNF4A, IGFBP2, CDKN2A/B, TCF7L2, KCNJ11, antioxidant genes, DNAJC3, PGC-1α, ADIPOQ, CDKAL1, POMC, PPARγ2, and SLC30A8.20–61
 |  |  |
Table 1 Association Between Genetic and Prediction of T2DM |
KLF14
The transcription factor, KLF14, is located on chromosome 7q32.3. Variations in this gene are associated with high-density lipoprotein (HDL)-cholesterol and T2DM.62,63 A previous study showed that KLF14 is involved in metabolism as a transcriptional activator as it regulates the gene networks that participate in lipid metabolism.64 KLF14 gene is assumed to be an ancient retrotransposed copy of KLF16 gene, presumably after the divergence between eutherians and marsupials65,66 due to its lack of introns and a high sequence homology with KLF16 gene. The maternal expression of KLF14 was associated with an increased risk of T2DM when carried on the maternal chromosome.67
The expression of KLF14 in adipose tissue was shown to be associated with a combined insulin resistance phenotype. It is characterized by increased fasting insulin and triglyceride levels and decreased HDL-cholesterol levels.68 Higher fasting insulin levels are manifested in the risk allele of rs4731702,67 such that the risk allele of this non-coding genetic variant could play a role in insulin resistance. Furthermore, it may act to influence the expression of genes associated with the body mass index (BMI) and the homeostasis model assessment for insulin resistance (HOMA-IR) due to its primary effects on insulin sensitivity, fasting glucose, and adiponectin.69 Moreover, rs4731702 was reported to be associated with gene expression in subcutaneous adipose tissue biopsies.68 Hence, it was suggested that KLF14 is the master transregulator of adipose tissue gene expression.70 One study also revealed that the G allele of KLF14 (rs972283) contributes to elevated blood pressure. Therefore, patients with metabolic syndrome have a greater risk of cardiovascular disease.20
KCNQ1
The KCNQ1 gene, which encodes the alpha-subunit of voltage-gated potassium channel Kv7.1, is a member of the Kv channel superfamily, and is located on chromosome 11p15.5.71,72 The protein that KCNQ1 gene encodes is the pore-forming alpha subunit of KCNQ1/KCNE1, KCNQ1/KCNE2 and KCNQ1/KCNE3 potassium channels.73 The expression of KvLQT1 repolarizes the action potential in cardiac muscles.20 KCNQ1 is also expressed in other tissues such as adipose tissue, the pancreas, and the brain.74
Mutations in KCN genes are associated with the development of diabetes. Variants in the KCNQ1 gene have been associated with reduced depolarization-evoked insulin exocytosis.24 The variant allele (C allele) of the rs2283228 [an intron variant according to National Center for Biotechnology Information (NCBI) dbSNP database: https://www.ncbi.nlm.nih.gov/snp/rs2283228] was shown to be associated with increased fasting glucose levels and impaired β-cell function in Asians.75 Moreover, previous studies showed that a vast majority of the genomic loci detected to date were associated with β-cell dysfunction in patients with T2DM.76–78 KCNQ1 is expressed in pancreatic islets and plays an essential role in glucose homeostasis as it functions as a regulator of insulin secretion.71,72 The KCNQ1 protein was shown to be expressed in insulin-secreting INS-1 cells.79 A study showed that the C allele of the intronic rs2237895 in KCNQ1 was associated with a decreased risk of abdominal obesity in patients with T2DM. These findings indicated that the C allele of rs2237895 is correlated with a decreased BMI and waist circumference in a Chinese population.80–82
DUSP9
DUSP9 encodes dual-specificity phosphatase 9 [also known as map kinase phosphatase 4 (MKP4)], mapped on chromosome X, with a cytogenetic location at Xq28. It is expressed in various tissues such as adipose tissue, muscles, insulin-responsive tissues, and the liver. DUSP9 plays important roles in regulating cell cycle and insulin action, and also has protective effects against the development of insulin resistance due to its ability to inactivate extracellular signal-regulated kinase and c-Jun N-terminal kinase. Therefore, DUSP9 was considered as a stress-induced insulin resistance mediator.83,84 While the effects of DUSP9 on insulin metabolism may differ depending on conditions and tissues, it is considered an important regulator of insulin sensitivity.25
The study of Voight et al62 first discovered an association of DUSP9 rs5945326 and T2DM risk in population of European descent. Then, the study of Fukuda et al25 replicated such an association in a Japanese population. A study of Rees et al26 showed that SNPs in or near DUSP9 and 12 other genomic loci showed significant associations with T2DM in Pakistani populations, with similar effect sizes to those seen in European populations.
FTO
Biological function of FTO (fat-mass and obesity associated) modulates the gene expression through methylation–demethylation modification since FTO is part of Fe(II)- and 2-oxoglutarate-dependent dioxygenases superfamily. Therefore, ubiquitously expressed hepatic FTO showed an important role in the homeostasis of glucose and lipid.85–89
Many studies have demonstrated a strong association between the FTO gene and the incidence of obesity, which is a major risk factor for T2DM.90–96 The majority of people with T2DM, particularly those of East Asian ethnicities, achieve their maximum lifetime BMI (BMImax) at the time of or before the onset of disease, and after T2DM diagnosis. The BMImax may also be reached after lifestyle interventions such as diet and exercise, and/or treatment with various antidiabetic medicines that may affect their obesity-related measurements, such as the BMI.97,98
A previous study has reported that the BMImax was strongly associated with an increased risk of T2DM. FTO SNPs were significantly correlated with the BMImax in a sex-stratified analysis.28 The study also found that rs1558902 was correlated with the incidence of T2DM in humans, and the correlations between SNPs and T2DM remained significant after the adjustment for the current age and BMI. Furthermore, Hertel et al also reported that adjusting the FTO variant for the waist-to-hip ratio and waist circumference conferred an increased risk of T2DM.29 Decreased mitochondrial oxidative capacities, oxidative stress, and lipid accumulation are suggested to increase the expression of FTO in patients with T2DM. Furthermore, the rs9939609 SNP may alter the risk of T2DM independent of the BMI by affecting other genes in the region.99 The increased FTO expression can stimulate de novo lipogenesis, inhibit lipolysis and fatty acid oxidation, and increase gluconeogenesis, which can lead to abnormally increased triglyceride deposition and the production of hepatic glucose (Figure 2).
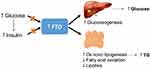 |
Figure 2 Impairments in the regulation of insulin and glucose may cause an increase in hepatic FTO expression. Abbreviation: TG, triglyceride. |
HNF4A
The HNF4A gene is a member of the steroid hormone receptor superfamily that is mainly expressed in the kidney, liver, pancreas (including β-cells), and small intestine, and influences metabolism and lipid transport.100,101 It also plays roles in liver function and hepatocyte differentiation.102,103 The HNF4A gene is composed of 13 exons and two promoters, known as P1 and P2.104 The P1 promoter is active mainly in liver cells,104–106 while the P2 promoter is the major splice variant in pancreatic β-cells.107−109
Approximately 1–2% of all diabetes cases are the monogenic form, known as maturity-onset diabetes of the young (MODY).110 It is characterized by an early age of onset (usually during adolescence or childhood), dominant inheritance, and defects in β-cell function. MODY resulting from mutations in the HNF4A transcription factor are known as MODY1.111 Studies on the genetic linkage have demonstrated that MODY1 is closely related to markers near HNF4A on chromosome 20.112
The non-coding variants of HNF4A gene rs6017317113 and rs481282936 and a coding missense variant rs1800961 (T130I)114 have been shown to play a role in the development of T2DM. In pancreatic β-cells (Figure 3), HNF4A is required for glucose metabolism and the expression and secretion of the normal insulin gene,115 while in the liver, HNF4A is required for hepatic gluconeogenesis.116 Yamagata et al screened for mutations in HNF4A in patients with MODY1 and reported that MODY1 is encoded by HNF4A.111 Clinical studies reported that MODY1 can be caused by impaired insulin secretion by pancreatic β-cells. Loss of or decreased HNF4A can lead to β-cell dysfunction.117 Based on these findings, HNF4A may participate in insulin secretion disorders, as seen in patients with T2DM and MODY1.
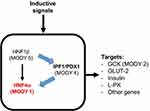 |
Figure 3 Transcription factor network in the pancreatic β-cell. Abbreviations: GCK, glucokinase; GLUT-2, glucose transporter-2; L-PK, liver pyruvate kinase. |
IGF2BP2
IGF2BP2 (insulin-like growth factor 2 mRNA-binding protein 2) was identified as an important T2DM candidate gene.31,118,119 It is located on chromosome 3q27 (https://www.genecards.org/cgi-bin/carddisp.pl?gene=IGF2BP2), and is highly expressed in pancreatic islet cells.31 In adipose tissue and the pancreas, IGF2BP2 can reduce the expression of IGF2, which is a growth factor that plays a crucial role in controlling pancreatic development and adipogenesis.120,121 IGF2BP2 plays roles in normal embryonic growth and development.122 It also plays a role in T2DM, which is associated with decreased insulin secretion.123 Hence, IGF2BP2 may support T2DM development via changes in adipose tissue or impaired β-cell function.
Duesing et al conducted a comprehensive genetic association study on French Caucasians and showed that IGF2BP2 rs4402960 and rs1470579 were associated with T2DM susceptibility.124 Another study reported higher levels of fasting plasma glucose, total cholesterol, and postprandial serum insulin in patients with T2DM who carried the C allele of rs1470579 compared with patients with T2DM who were AA carriers. IGF2BP2 polymorphisms play a role in the regulation of pancreatic β-cell function.125 Studies have also demonstrated that IGF2BP2 is strongly associated with overweight and obesity.38 Obesity is associated with T2DM; hence, it is hypothesized that the association between IGF2BP2 and T2DM may be modified by obesity. This is also known as the interplay between IGF2BP2 and obesity with T2DM.126 In keeping with this hypothesis, Chistiakov and co-workers,127 reported that patients with T2DM have a more than twofold increase in IGF2BP2 expression levels in adipose tissue compared with healthy individuals. Associations between IGF2BP2 and visceral/abdominal total fat were also demonstrated in Mexican Americans and Canadian Caucasians, proposing a possible role of IGF2BP2 in insulin resistance.128
CDKN2A/B
The CDKN2A/B locus is located on chromosome 9p21.3, such that the CDKN2A gene encodes both the p16 inhibitor of cyclin-dependent kinase p16INK4A and p14ARF, and the CDKN2B gene encodes p15INK4B,129 respectively, and this locus has been associated with T2DM risk.43,118 Further, the 9p21 SNP rs10811661, which was associated with the expression of a long non-coding RNA known as antisense noncoding RNA in the INK4 locus [ANRIL; also called CDKN2B antisense RNA 1 (CDKN2B-AS1)],130 was linked with the risk of human diabetes in a GWAS.118 Polymorphisms in CDKN2A/B affect metabolic health related to proteins that contribute to the regulation of β-cell mass, insulin secretory function, and proliferation.43 Additional studies in Asia and Europe have also confirmed that CDKN2A/B is associated with T2DM risk.40–42,44,56,131–133 CDKN2A/B is highly expressed in adipocytes and islet cells, as well as in brain cells. Both CDKN2A and CDKN2B are tumor suppressor genes involved in cell apoptosis, tumorigenesis, and proliferation.134
Alterations to the phenotype of immune cells influence systemic and peripheral insulin resistance and lead to T2DM. Especially in obesity condition, macrophage infiltrates into adipose tissue and lead to develop a chronic low-grade inflammation. These adipose tissue macrophages (ATMs) stimulate pro-inflammatory cytokines secretion and further will contribute to insulin resistance.135 Additionally, CDKN2A/B-ANRIL gene products control glucose homeostasis, in part, via the control of insulin secretion and β-cell function (Figure 4).
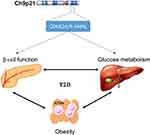 |
Figure 4 Probable mechanism of CKN2A/B-antisense noncoding RNA in the INK4 locus (ANRIL) gene product. Abbreviation: ATM, adipose tissue macrophage. |
TCF7L2
TCF7L2 (transcription factor 7-like 2) is a transcription factor that plays a role in the Wnt-signaling pathway, which regulates pancreatic islet cell functions, such as proliferation and cell survival.136 A previous study showed that increased β-cell apoptosis was associated with decreased TCF7L2 activity, resulting in the downregulation of insulin secretion.137,138
The TCF7L2 gene is located on chromosome 10q.25.2–25.3, also known as the TCF4 locus. Previous studies have indicated that people with T2DM are more likely to carry the genetic variant (rs7903146) of this gene.139–141 Furthermore, studies on various ethnic populations have shown that mutations of this gene are associated with TCF7L2 in a self-regulating manner via transcriptional protein complex binding across rs7903146.142–144
The Wnt-signaling pathway also controls the transcription of the proglucagon gene, which regulates incretin hormones such as glucagon-like peptide-1 that inhibits glucagon activity and maintains food mobility from the stomach to the duodenum, and gastric inhibitory polypeptide that is produced by intestinal K cells. Mutations in TCF7L2 also result in reduced expression of the proglucagon gene and, consequently, reduced glucagon-like peptide-1 production.145–147
TCF7L2 is expressed in other organs, such as skeletal muscle, gut, fat, and liver, which are all also involved in mediating metabolic homeostasis.148 The overexpression of β-catalase produced reciprocal effects on hepatic gluconeogenesis.149 On the other hand, the Wnt-signaling pathway negatively regulates adipogenesis, and Wnt ligands produced by adipocytes may also function as endocrine and paracrine factors.150 Based on those studies, the possible roles of TCF7L2 in the pathogenesis of T2DM are summarized in Figure 5.
 |
Figure 5 Possible role of TCF7L2 in the pathogenesis of T2DM. Abbreviations: GIP, gastric inhibitory polypeptide; GLP-1, glucagon-like peptide 1. |
KCNJ11
The KCNJ11 gene (potassium channel, inwardly rectifying, subfamily J, member 11) encodes the Kir6.2 protein (inward-rectifier potassium ion channel), which is important for insulin secretion via the ATP-sensitive potassium (KATP) channel. It has no intron region and is located on chromosome 11p15.1.151 As described in Figure 6, when the body demands insulin, Kir6.2 couples itself to SUR1 (sulfonylurea receptor-1) and binds to a KATP channel on the pancreatic β-cell membrane, leading to insulin production. Increased glucose levels stimulate the KATP channel to open and allow the entry of K+ ions. Increasing levels of K+ ions depolarize the cell membrane and induce Ca2+ channels to increase levels of free intracellular Ca2+. The Ca2+ ions trigger other components of the insulin secretion pathway to release granules.152,153 Therefore, mutations in KCNJ11 result in reduced insulin production due to reduced or absent Kir6.2 protein expression.154 The variant allele of KCNJ11 gene rs5219 may decrease channel sensitivity to ATP and alter the charge of the ATP-binding region.51 A recent meta-analysis showed a strong relationship between polymorphisms of rs5219 and susceptibility to T2DM in East Asian and Caucasian populations.155 Kir6.2 is also expressed in neurons, the brain, and muscles.156
Antioxidant Genes
Disruption to the balance of antioxidants and reactive oxygen species (ROS) results in increased oxidative stress, which may lead to diabetes. The generation and accumulation of ROS in β-cells can cause β-cell dysfunction, defects in insulin production, and impaired function, which result in diabetes.157 However, the impact of oxidative stress can be reduced or modified by enzymatic antioxidants, including catalase (CAT), glutathione-S-transferase (GST), glutathione peroxidase (GPx), superoxide dismutase (SOD), nitric oxide synthase, and nicotinamide adenine dinucleotide phosphate oxidase.158–160 Banerjee et al reported that individuals with a polymorphism affecting the genetic regulation of these six enzymes were at increased risk of developing T2DM. Known polymorphisms in these genes include GSTM1del, GSTT1del, GSTP1 105I/V(+313A/G), CAT-21A/T, SOD2 + 47C/T, and GPx1 + 599C/T.161 Banerjee and co-workers also concluded that the risk of developing T2DM increases as the variation of the genes that regulate antioxidant enzyme increases.161
DNAJC3
As explained by DNAJC3 is an endoplasmic reticulum (ER) lumen protein and a member of the HSP70 family. It is located in all tissues in humans (predominantly the liver and pancreas), and plays a role in maintaining homeostasis in the ER.54 It serves as co-chaperone of binding immunoglobulin protein (BiP) during the unfolded protein response (UPR), which is an ER adaptive signaling pathway. Normally, the ER regulates membrane homeostasis by synthesizing and modifying secretory and membrane proteins.54 However, when cells are exposed to abnormal conditions, such as infection, homeostasis imbalance, glucose deprivation, or stimulation that leads to ER protein overproduction, the proteins undergo incomplete or abnormal processes that form unfolded or misfolded proteins. The accumulation of these proteins increases stress in the ER lumen, eventually triggering the UPR in the ER.54
Three pathways were reported to generate the UPR signaling pathway, including activation of transcription factor-6;162 activation of inositol-requiring transmembrane kinase/endoribonuclease 1;163 and double-stranded RNA-dependent protein kinase-like eukaryotic initiation factor 2α kinase (PERK).164
The UPR pathways will reduce the ER stress and maintain the cell survival by correcting the misinterpreted protein. This can be carried out by the SIL1 protein, which interacts with BiP and binds the misinterpreted protein. DNAJC3 acts prior to protein correction. It binds reversibly to hydrophobic segments of the protein and delivers it to the chaperone, BiP.54 DNAJC3 is involved in the PERK pathway, collaborating with the chaperone, BiP, and SIL1 protein, a nucleotide exchange factor.54 DNAJC3 mutations, such as deletions and stop mutations, result in reduced or absent binding between BiP and unfolded or misfolded proteins.54 In summary, adaptive response failure leads to unsuccessful pancreatic ER homeostasis and cell death; and in pancreatic cell death, particularly in the pancreatic islet, this will reduce insulin production. Therefore, mutations in the DNAJC3 gene are correlated with diabetes.54
PGC-1α
PGC-1α (peroxisome proliferator-activated receptor (PPAR)-γ coactivator-1α) is a transcription coactivator that is involved in various biological responses, such as temperature adaptation, energy homeostasis in the mitochondria, glucose metabolism, triglyceride homeostasis, and heart development.165
In humans, the PPARGC1A gene is located on chromosome 4 and encodes a 798-amino acid protein PGC-1α, which is expressed in most tissues with highly active mitochondria and oxidative metabolism, such as the heart, skeletal muscle, and brown adipose tissue.166
The PGC-1α gene can convert muscle fiber type and stimulate fatty acid oxidation, thus may lead to a decreased fatty acid concentration in muscles.167 In contrast, a recent study showed that insulin-resistant skeletal muscle and the liver were associated with increased levels of triglycerides.168
A previous study by Kelley and co-workers suggested that a decrease in mitochondrial oxidative enzymes leads to defects in the mitochondrial fatty acid oxidation pathway and, ultimately, diabetes. Furthermore, the study also observed that patients with T2DM showed downregulated expression of PPARGC1A gene.169
ADIPOQ
The ADIPOQ gene encodes adiponectin, which is an adipocytokine. The adipose tissue plays an important role in the development of diabetes mellitus and obesity.170 Adiponectin, a major adipocyte secretory protein in human plasma, functions as a regulator of energy and is involved in glucose tolerance.171 The ADIPOQ gene is located on human chromosome 3q27 and is reported to be a susceptibility locus for T2DM.172,173
Adiponectin is associated with increased insulin sensitivity, reduced ER stress, and increased fatty acid oxidation.174 Its functions are mediated by its receptors, AdipoR1 and AdipoR2. The binding of adiponectin to its receptor activates intracellular signaling molecules that play important roles in lipid and carbohydrate metabolism.175
Li et al reported ADIPOQ gene polymorphisms in rs1501299, rs182052, and rs7627128 in a Chinese population, and showed a significant association with T2DM. Furthermore, a haplotype-based case-control study on the association between T2DM and the ADIPOQ gene found that the haplotypes A–A–T and G–A–T were correlated with increased potency and decreased risk of T2DM, respectively.55
CDKAL1
Klimentidis et al reported that variations in CDKAL1 rs775480 were associated with hemoglobin A1c, which is related to T2DM. The rs775480 polymorphism is located at intron 5 of the CDKAL1 gene.57 This SNP is associated with decreased glucose sensitivity and insulin secretion in β-cells.176,177 Furthermore, the rs10946398 polymorphism of the CDKAL1 gene was proposed as a marker of impaired insulin secretion, as the CC/CA genotypes and C allele contribute to T2DM susceptibility in obese individuals.56,178,179
POMC
Mutations in the POMC (pro-opiomelanocortin) gene are reportedly associated with overweight and obesity as well as the phenotype of early-onset T2DM.58,180,181 POMC is a precursor polypeptide hormone that is produced in the neurons of the arcuate nucleus of the hypothalamus and plays an important role as a controller of homeostasis, as well as energy balance, food intake, and glucose metabolism.182–184
Mencarelli et al reported that patients with T2DM and obesity related to mutations in the POMC genes showed a missense mutation in the signal peptide.58 This mutation led to a heterozygous substitution of arginine for glycine at A15G–POMC (codon 15), which inhibited the production and secretion of the POMC protein. In humans, POMC deficiency can cause insulin resistance (hyperinsulinemia) since POMC-derived peptides have local effects on the central melanocortin pathway, and intact neuronal melanocortin signaling regulates insulin sensitivity in peripheral tissues.185,186
PPARγ2
PPARγ2 (peroxisome proliferator-activated receptor-gamma 2) is a ligand-activated transcription factor of the nuclear hormone receptor superfamily.187 The PPARγ2 gene plays roles in glucose homeostasis, lipid metabolism, obesity, insulin sensitivity, T2DM, and various adipocyte-specific genes.59,188–190
Based on several case-control and family-based studies, estimated that Pro12 allele (ie, the major allele) of PPARγ was associated with a 1.25-fold elevated risk of T2DM.191 Further, the study of Chan et al showed that the Pro12Ala polymorphism was associated with T2DM risk in the multiethnic Women’s Health Initiative (WHI) Observational Study at a nominal significance level (Pro12 allele is the risk-increasing allele, p=0.01, additive model). The study was replicated in the WHI SNP Health Association Resource (WHI-SHARe) Hispanic American case-control sample (Pro12 allele is the risk-increasing allele, p=0.02, additive model).192
Phani et al reported that the PPARγ2 gene was associated with T2DM in an obese diabetic Indian population (BMI ≥ 25 kg/m2).193 The rs1801282 polymorphism in the PPARγ2 gene has been associated with adiposity and regulation of the BMI. Furthermore, the Ala12 variant allele of rs1801282 has been shown to exhibit a decreased binding affinity to the cognate DNA element and therefore could reduce PPARγ2 transcriptional activity.194 Based on the study of Valve et al, the Ala12 variant allele was associated with a lower BMI and a higher insulin sensitivity among normal weight and mildly obese individuals.195 This polymorphism has also been linked to increased insulin sensitivity and protects from T2DM in Caucasian populations.59,193
Motavallian et al compared the allele distributions of Pro12Ala polymorphism between healthy individuals and those with diabetes.188 They found a higher frequency of the Ala allele12 in healthy individuals than in patients with diabetes. Another study found a protective role of high Ala frequency against T2DM as it was associated with increased insulin sensitivity, while low frequency of the Ala12 allele was associated with decreased insulin sensitivity (insulin resistance), which may lead to diabetes. These findings suggest that polymorphisms in the PPARγ2 gene are associated with T2DM.
SLC30A8
Previous studies have reported that the SLC30A8 (solute carrier family 30 member 8) rs13266634 polymorphism in the major C allele was strongly associated with the risk of T2DM.60,61 In addition, Chang et al also reported that the SLC30A8 rs13266634 SNP was associated with age as a T2DM risk factor.196
SLC30A8 is expressed in pancreatic β-cells and encodes a zinc transporter.196 Zinc is an important element for insulin secretion and storage.197 Low ZnT8 (zinc transporter-protein member 8) expression leads to decreased insulin production by β-cells. Low Zn2+ production facilitates hormone clearance by the liver (Figure 7). The study using ZnT8KO mice had low peripheral blood insulin levels despite hypersecretion from β cells pancreas, whilst reduced Zn2+ production favors clearance of the hormone by liver.32 Furthermore, ZnT8 overexpression increasing Zn2+ accumulation, the Zn2+ that secreted with insulin suppressed hepatic insulin clearance via the inhibition of clathrin-dependent insulin endocytosis.32,198 The SLC30A8 gene encodes ZnT8, which forms a solid hexamer from binding with insulin in β-cells, matures, and is stored in secretory vesicles.199
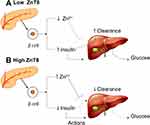 |
Figure 7 Interaction between ZnT8 expression (A) low ZnT8 and (B) high ZnT8, hormone action, and hepatic insulin clearance. |
Zn2+ plays a crucial role in insulin release and regulates the homeostasis of insulin concentration between pancreas and body. When blood glucose level is low, Zn2+ binds insulin in pancreas for storage purpose and an increase in blood glucose level will liberate insulin from Zn2+ High level of ZnT8 means there is a lot of Zn2+ available for bind and hold insulin in pancreas. In other words, insulin secretion will be limited to an increase in blood glucose and this is a normal physiology of body in maintaining the glucose homeostasis. Besides, a low level of ZnT8 indicates a small concentration of Zn2+ which means there is inadequate insulin depositor and thus, insulin hypersecretion will occur.200 The hypersecretion of insulin will impact the insulin sensitivity, liver clearance, and blood glucose level as the following statement, first liver will intoxicate an excessive amount of insulin. In other words, hepatic clearance will increase and liver takes more energy to function, resulting in glycogen breakdown to glucose.201 Second, Zn2+ also presents in insulin targeted cells to improve the sensitivity for insulin-receptor bind. A lack of Zn2+ will reduce the insulin sensitivity and decrease insulin-receptor bind affinity.202 These mechanisms will lead to an increase in blood glucose level or T2DM event.
The SLC30A8 rs13266634 polymorphism is a non-synonymous SNP that causes an amino acid change from arginine, encoded by the C-allele, to tryptophan, encoded by the T-allele, at position 325 (Arg325Trp). This polymorphism has been linked with the development of T2DM in several populations.31,118,119,139,140,203,204
T2DM risk is influenced by both genetic and environmental risk factors. Therefore, gene–environment interaction studies in T2DM could be more explored as indicated by other studies showed that a significant interaction between SLC30A8 gene rs13266634 and age in T2DM risk (p<0.0001).196,205
Conclusion and Future Prospects
Some genetic polymorphisms are associated with T2DM, either in the form of regulatory non-coding SNPs or as missense coding SNPs that cause direct changes to amino acids within a protein. Genes that are considered to predict or be associated with T2DM disrupt homeostasis, including insulin action and sensitivity, β-cell function and proliferation, and obesity. We realized that this review might use an incomplete searching method and some relevant papers have not been included, but it summarized genes that might be related to the development of T2DM. Moreover, studies show that different SNPs and mechanisms lead to diabetes in different ethnic groups.
Despite remarkable progress, the results from these genetic studies remain inconclusive. Therefore, future studies are required using different ethnic groups to confirm these findings globally, to determine correlations between gene expression and the mechanisms involved to confirm the suggested pathways, and to ensure that treatment of a specific gene will not have knock-on adverse effects on other genes. Thus, further intensive studies are necessary to identify more T2DM-associated genes. The evaluation and confirmation of the currently identified genes are also necessary due to conflicting findings. These polymorphisms may help to reduce the incidence and predict the risk of T2DM. Early identification may increase the prevention efficacy and increase prediabetic prognosis significantly.
Acknowledgment
This research is partially funded by the Indonesian Ministry of Research, Technology, and Higher Education of Republic of Indonesia under WCU program managed by Institute Teknologi Bandung for RA.
Disclosure
All authors declare that there is no conflict of interest related to this study.
References
1. Carrera Boada CA, Martinez-Moreno JM. Pathophysiology of diabetes mellitus type 2: beyond the duo “insulin resistance-secretion deficit”. Nutr Hosp. 2013;28(Suppl 2):78–87. doi:10.3305/nh.2013.28.sup2.6717
2. Danaei G, Finucane MM, Lu Y, et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet. 2011;378(9785):31–40. doi:10.1016/S0140-6736(11)60679-X
3. Shaw JE, Sicree RA, Zimmet PZ. Global estimates of the prevalence of diabetes for 2010 and 2030. Diabetes Res Clin Pract. 2010;87(1):4–14. doi:10.1016/j.diabres.2009.10.007
4. O’Rahilly S, Barroso I, Wareham NJ. Genetic factors in type 2 diabetes: the end of the beginning? Science. 2005;307(5708):370–373. doi:10.1126/science.1104346
5. Prokopenko I, McCarthy MI, Lindgren CM. Type 2 diabetes: new genes, new understanding. Trends Genet. 2008;24(12):613–621. doi:10.1016/j.tig.2008.09.004
6. Mahajan A, Taliun D, Thurner M, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50(11):1505–1513. doi:10.1038/s41588-018-0241-6
7. McCarthy MI, Zeggini E. Genome-wide association studies in type 2 diabetes. Curr Diab Rep. 2009;9(2):164–171. doi:10.1007/s11892-009-0027-4
8. Wheeler E, Barroso I. Genome-wide association studies and type 2 diabetes. Brief Funct Genomics. 2011;10(2):52–60. doi:10.1093/bfgp/elr008
9. Zheng JS, Li K, Huang T, et al. Genetic risk score of nine type 2 diabetes risk variants that interact with erythrocyte phospholipid alpha-linolenic acid for type 2 diabetes in Chinese hans: a case-control study. Nutrients. 2017;9(4):376. doi:10.3390/nu9040376
10. Lall K, Magi R, Morris A, Metspalu A, Fischer K. Personalized risk prediction for type 2 diabetes: the potential of genetic risk scores. Genet Med. 2017;19(3):322–329. doi:10.1038/gim.2016.103
11. Kong X, Xing X, Zhang X, Hong J, Yang W. Early-onset type 2 diabetes is associated with genetic variants of beta-cell function in the Chinese Han population. Diabetes Metab Res Rev. 2019;e3214.
12. Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. N Engl J Med. 2008;359(21):2208–2219. doi:10.1056/NEJMoa0804742
13. van Hoek M, Dehghan A, Witteman JC, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes. 2008;57(11):3122–3128. doi:10.2337/db08-0425
14. Cornelis MC, Qi L, Zhang C, et al. Joint effects of common genetic variants on the risk for type 2 diabetes in U.S. men and women of European ancestry. Ann Intern Med. 2009;150(8):541–550. doi:10.7326/0003-4819-150-8-200904210-00008
15. Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. N Engl J Med. 2008;359(21):2220–2232. doi:10.1056/NEJMoa0801869
16. Groop L, Forsblom C, Lehtovirta M, et al. Metabolic consequences of a family history of NIDDM (the Botnia study): evidence for sex-specific parental effects. Diabetes. 1996;45(11):1585–1593. doi:10.2337/diab.45.11.1585
17. Herder C, Kowall B, Tabak AG, Rathmann W. The potential of novel biomarkers to improve risk prediction of type 2 diabetes. Diabetologia. 2014;57(1):16–29. doi:10.1007/s00125-013-3061-3
18. Abuissa H, Bel DS, J H O
19. Schwingshackl L, Hoffmann G, Lampousi AM, et al. Food groups and risk of type 2 diabetes mellitus: a systematic review and meta-analysis of prospective studies. Eur J Epidemiol. 2017;32(5):363–375. doi:10.1007/s10654-017-0246-y
20. Kong X, Zhang X, Xing X, Zhang B, Hong J, Yang W. The association of type 2 diabetes loci identified in genome-wide association studies with metabolic syndrome and its components in a chinese population with type 2 diabetes. PLoS One. 2015;10(11):e0143607. doi:10.1371/journal.pone.0143607
21. Zyriax BC, Salazar R, Hoeppner W, Vettorazzi E, Herder C, Windler E. The association of genetic markers for type 2 diabetes with prediabetic status – cross-sectional data of a diabetes prevention trial. PLoS One. 2013;8(9):e75807. doi:10.1371/journal.pone.0075807
22. Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–1097. doi:10.1038/ng.207
23. Stancakova A, Kuulasmaa T, Paananen J, et al. Association of 18 confirmed susceptibility loci for type 2 diabetes with indices of insulin release, proinsulin conversion, and insulin sensitivity in 5,327 nondiabetic Finnish men. Diabetes. 2009;58(9):2129–2136. doi:10.2337/db09-0117
24. Khan IA, Vattam KK, Jahan P, Mukkavali KK, Hasan Q, Rao P. Correlation between KCNQ1 and KCNJ11 gene polymorphisms and type 2 and post-transplant diabetes mellitus in the Asian Indian population. Genes Dis. 2015;2(3):276–282. doi:10.1016/j.gendis.2015.02.009
25. Fukuda H, Imamura M, Tanaka Y, et al. A single nucleotide polymorphism within DUSP9 is associated with susceptibility to type 2 diabetes in a Japanese population. PLoS One. 2012;7(9):e46263. doi:10.1371/journal.pone.0046263
26. Rees SD, Hydrie MZ, Shera AS, et al. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia. 2011;54(6):1368–1374. doi:10.1007/s00125-011-2063-2
27. Bao XY, Peng B, Yang MS. Replication study of novel risk variants in six genes with type 2 diabetes and related quantitative traits in the Han Chinese lean individuals. Mol Biol Rep. 2012;39(3):2447–2454. doi:10.1007/s11033-011-0995-8
28. Kamura Y, Iwata M, Maeda S, et al. FTO gene polymorphism is associated with type 2 diabetes through its effect on increasing the maximum BMI in Japanese men. PLoS One. 2016;11(11):e0165523. doi:10.1371/journal.pone.0165523
29. Hertel JK, Johansson S, Sonestedt E, et al. FTO, type 2 diabetes, and weight gain throughout adult life: a meta-analysis of 41,504 subjects from the Scandinavian HUNT, MDC, and MPP studies. Diabetes. 2011;60(5):1637–1644. doi:10.2337/db10-1340
30. Ortega-Azorin C, Sorli JV, Asensio EM, et al. Associations of the FTO rs9939609 and the MC4R rs17782313 polymorphisms with type 2 diabetes are modulated by diet, being higher when adherence to the Mediterranean diet pattern is low. Cardiovasc Diabetol. 2012;11:137. doi:10.1186/1475-2840-11-137
31. Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi:10.1126/science.1142382
32. Tamaki M, Fujitani Y, Hara A, et al. The diabetes-susceptible gene SLC30A8/ZnT8 regulates hepatic insulin clearance. J Clin Invest. 2013;123(10):4513–4524. doi:10.1172/JCI68807
33. Mtiraoui N, Turki A, Nemr R, et al. Contribution of common variants of ENPP1, IGF2BP2, KCNJ11, MLXIPL, PPARgamma, SLC30A8 and TCF7L2 to the risk of type 2 diabetes in Lebanese and Tunisian Arabs. Diabetes Metab. 2012;38(5):444–449. doi:10.1016/j.diabet.2012.05.002
34. Bonnycastle LL, Willer CJ, Conneely KN, et al. Common variants in maturity-onset diabetes of the young genes contribute to risk of type 2 diabetes in Finns. Diabetes. 2006;55(9):2534–2540. doi:10.2337/db06-0178
35. Marcil V, Amre D, Seidman EG, et al. Hepatocyte nuclear factor 4 alpha polymorphisms and the metabolic syndrome in French-Canadian youth. PLoS One. 2015;10(2):e0117238. doi:10.1371/journal.pone.0117238
36. Kooner JS, Saleheen D, Sim X, et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984–989. doi:10.1038/ng.921
37. Rao P, Wang H, Fang H, et al. Association between IGF2BP2 polymorphisms and type 2 diabetes mellitus: a case-control study and meta-analysis. Int J Environ Res Public Health. 2016;13:6. doi:10.3390/ijerph13060574
38. Lasram K, Ben Halim N, Benrahma H, et al. Contribution of CDKAL1 rs7756992 and IGF2BP2 rs4402960 polymorphisms in type 2 diabetes, diabetic complications, obesity risk and hypertension in the Tunisian population. J Diabetes. 2015;7(1):102–113. doi:10.1111/1753-0407.12147
39. Chauhan G, Spurgeon CJ, Tabassum R, et al. Impact of common variants of PPARG, KCNJ11, TCF7L2, SLC30A8, HHEX, CDKN2A, IGF2BP2, and CDKAL1 on the risk of type 2 diabetes in 5,164 Indians. Diabetes. 2010;59(8):2068–2074. doi:10.2337/db09-1386
40. Gamboa-Melendez MA, Huerta-Chagoya A, Moreno-Macias H, et al. Contribution of common genetic variation to the risk of type 2 diabetes in the Mexican Mestizo population. Diabetes. 2012;61(12):3314–3321. doi:10.2337/db11-0550
41. Chen G, Xu Y, Lin Y, et al. Association study of genetic variants of 17 diabetes-related genes/loci and cardiovascular risk and diabetic nephropathy in the Chinese She population. J Diabetes. 2013;5(2):136–145. doi:10.1111/1753-0407.12025
42. Hu C, Zhang R, Wang C, et al. PPARG, KCNJ11, CDKAL1, CDKN2A-CDKN2B, IDE-KIF11-HHEX, IGF2BP2 and SLC30A8 are associated with type 2 diabetes in a Chinese population. PLoS One. 2009;4(10):e7643. doi:10.1371/journal.pone.0007643
43. Xiao S, Zeng X, Fan Y, et al. Gene polymorphism association with type 2 diabetes and related gene–gene and gene–environment interactions in a Uyghur Population. Med Sci Monit. 2016;22:474–487. doi:10.12659/msm.895347
44. Wen J, Ronn T, Olsson A, et al. Investigation of type 2 diabetes risk alleles support CDKN2A/B, CDKAL1, and TCF7L2 as susceptibility genes in a Han Chinese cohort. PLoS One. 2010;5(2):e9153. doi:10.1371/journal.pone.0009153
45. Corella D, Coltell O, Sorli JV, et al. Polymorphism of the transcription factor 7-like 2 gene (TCF7L2) interacts with obesity on type-2 diabetes in the PREDIMED study emphasizing the heterogeneity of genetic variants in type-2 diabetes risk prediction: time for obesity-specific genetic risk scores. Nutrients. 2016;8(12):793.
46. Cauchi S, Meyre D, Dina C, et al. Transcription factor TCF7L2 genetic study in the French population: expression in human beta-cells and adipose tissue and strong association with type 2 diabetes. Diabetes. 2006;55(10):2903–2908. doi:10.2337/db06-0474
47. Sale MM, Smith SG, Mychaleckyj JC, et al. Variants of the transcription factor 7-like 2 (TCF7L2) gene are associated with type 2 diabetes in an African-American population enriched for nephropathy. Diabetes. 2007;56(10):2638–2642. doi:10.2337/db07-0012
48. Ciccacci C, Di Fusco D, Cacciotti L, et al. TCF7L2 gene polymorphisms and type 2 diabetes: association with diabetic retinopathy and cardiovascular autonomic neuropathy. Acta Diabetol. 2013;50(5):789–799. doi:10.1007/s00592-012-0418-x
49. Wang J, Zhang J, Li L, et al. Association of rs12255372 in the TCF7L2 gene with type 2 diabetes mellitus: a meta-analysis. Braz J Med Biol Res. 2013;46(4):382–393. doi:10.1590/1414-431x20132677
50. Nanfa D, Sobngwi E, Atogho-Tiedeu B, et al. Association between the TCF7L2 rs12255372 (G/T) gene polymorphism and type 2 diabetes mellitus in a Cameroonian population: a pilot study. Clin Transl Med. 2015;4:17. doi:10.1186/s40169-015-0058-1
51. Liu Z, Zhang YW, Feng QP, et al. [Association analysis of 30 type 2 diabetes candidate genes in Chinese Han population]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2006;28(2):124–128.
52. Phani NM, Guddattu V, Bellampalli R, et al. Population specific impact of genetic variants in KCNJ11 gene to type 2 diabetes: a case-control and meta-analysis study. PLoS One. 2014;9(9):e107021. doi:10.1371/journal.pone.0107021
53. Abdelhamid I, Lasram K, Meiloud G, et al. E23K variant in KCNJ11 gene is associated with susceptibility to type 2 diabetes in the Mauritanian population. Prim Care Diabetes. 2014;8(2):171–175. doi:10.1016/j.pcd.2013.10.006
54. Synofzik M, Haack TB, Kopajtich R, et al. Absence of BiP co-chaperone DNAJC3 causes diabetes mellitus and multisystemic neurodegeneration. Am J Hum Genet. 2014;95(6):689–697. doi:10.1016/j.ajhg.2014.10.013
55. Li ZP, Zhang M, Gao J, Zhou GY, Li SQ, An ZM. Relation between ADIPOQ gene polymorphisms and type 2 diabetes. Genes. 2015;6(3):512–519. doi:10.3390/genes6030512
56. Han X, Luo Y, Ren Q, et al. Implication of genetic variants near SLC30A8, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, FTO, TCF2, KCNQ1, and WFS1 in type 2 diabetes in a Chinese population. BMC Med Genet. 2010;11:81. doi:10.1186/1471-2350-11-81
57. Klimentidis YC, Lemas DJ, Wiener HH, et al. CDKAL1 and HHEX are associated with type 2 diabetes-related traits among Yup’ik people. J Diabetes. 2014;6(3):251–259. doi:10.1111/1753-0407.12093
58. Mencarelli M, Zulian A, Cancello R, et al. A novel missense mutation in the signal peptide of the human POMC gene: a possible additional link between early-onset type 2 diabetes and obesity. Eur J Hum Genet. 2012;20(12):1290–1294. doi:10.1038/ejhg.2012.103
59. Deeb SS, Fajas L, Nemoto M, et al. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20(3):284–287. doi:10.1038/3099
60. Dong F, Zhang BH, Zheng SL, et al. Association between SLC30A8 rs13266634 polymorphism and risk of T2DM and IGR in Chinese population: a systematic review and meta-analysis. Front Endocrinol (Lausanne). 2018;9:564. doi:10.3389/fendo.2018.00564
61. Fan M, Li W, Wang L, et al. Association of SLC30A8 gene polymorphism with type 2 diabetes, evidence from 46 studies: a meta-analysis. Endocrine. 2016;53(2):381–394. doi:10.1007/s12020-016-0870-4
62. Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi:10.1038/ng.609
63. Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi:10.1038/nature09270
64. de Assuncao TM, Lomberk G, Cao S, et al. New role for Kruppel-like factor 14 as a transcriptional activator involved in the generation of signaling lipids. J Biol Chem. 2014;289(22):15798–15809. doi:10.1074/jbc.M113.544346
65. Parker-Katiraee L, Carson AR, Yamada T, et al. Identification of the imprinted KLF14 transcription factor undergoing human-specific accelerated evolution. PLoS Genet. 2007;3(5):e65. doi:10.1371/journal.pgen.0030065
66. Okamura K, Nakai K. Retrotransposition as a source of new promoters. Mol Biol Evol. 2008;25(6):1231–1238. doi:10.1093/molbev/msn071
67. Kong A, Steinthorsdottir V, Masson G, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868–874. doi:10.1038/nature08625
68. Small KS, Todorcevic M, Civelek M, et al. Regulatory variants at KLF14 influence type 2 diabetes risk via a female-specific effect on adipocyte size and body composition. Nat Genet. 2018;50(4):572–580. doi:10.1038/s41588-018-0088-x
69. Dimas AS, Lagou V, Barker A, et al. Impact of type 2 diabetes susceptibility variants on quantitative glycemic traits reveals mechanistic heterogeneity. Diabetes. 2014;63(6):2158–2171. doi:10.2337/db13-0949
70. Small KS, Hedman AK, Grundberg E, et al. Identification of an imprinted master trans regulator at the KLF14 locus related to multiple metabolic phenotypes. Nat Genet. 2011;43(6):561–564. doi:10.1038/ng1011-1040c
71. Yamagata K, Senokuchi T, Lu M, et al. Voltage-gated K+ channel KCNQ1 regulates insulin secretion in MIN6 beta-cell line. Biochem Biophys Res Commun. 2011;407(3):620–625. doi:10.1016/j.bbrc.2011.03.083
72. Mulder H, Nagorny CL, Lyssenko V, Groop L. Melatonin receptors in pancreatic islets: good morning to a novel type 2 diabetes gene. Diabetologia. 2009;52(7):1240–1249. doi:10.1007/s00125-009-1359-y
73. Chen YH, Xu SJ, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299(5604):251–254. doi:10.1126/science.1077771
74. Demolombe S, Franco D, de Boer P, et al. Differential expression of KvLQT1 and its regulator IsK in mouse epithelia. Am J Physiol Cell Physiol. 2001;280(2):C359–372. doi:10.1152/ajpcell.2001.280.2.C359
75. Tan JT, Nurbaya S, Gardner D, Ye S, Tai ES, Ng DP. Genetic variation in KCNQ1 associates with fasting glucose and beta-cell function: a study of 3,734 subjects comprising three ethnicities living in Singapore. Diabetes. 2009;58(6):1445–1449. doi:10.2337/db08-1138
76. Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi:10.1038/ng.120
77. Simonis-Bik AM, Nijpels G, van Haeften TW, et al. Gene variants in the novel type 2 diabetes loci CDC123/CAMK1D, THADA, ADAMTS9, BCL11A, and MTNR1B affect different aspects of pancreatic beta-cell function. Diabetes. 2010;59(1):293–301. doi:10.2337/db12-1627
78. Jonsson A, Ladenvall C, Ahluwalia TS, et al. Effects of common genetic variants associated with type 2 diabetes and glycemic traits on alpha- and beta-cell function and insulin action in humans. Diabetes. 2013;62(8):2978–2983. doi:10.2337/db12-1627
79. Ullrich S, Su J, Ranta F, et al. Effects of I(Ks) channel inhibitors in insulin-secreting INS-1 cells. Pflugers Arch. 2005;451(3):428–436. doi:10.1007/s00424-005-1479-2
80. Liu Y, Zhou DZ, Zhang D, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes in the population of mainland China. Diabetologia. 2009;52(7):1315–1321. doi:10.1007/s00125-009-1375-y
81. Yu W, Ma RC, Hu C, et al. Association between KCNQ1 genetic variants and obesity in Chinese patients with type 2 diabetes. Diabetologia. 2012;55(10):2655–2659. doi:10.1007/s00125-012-2636-8
82. Kong X, Hong J, Chen Y, et al. Association of genetic variants with isolated fasting hyperglycaemia and isolated postprandial hyperglycaemia in a Han Chinese population. PLoS One. 2013;8(8):e71399. doi:10.1371/journal.pone.0071399
83. Xu H, Dembski M, Yang Q, et al. Dual specificity mitogen-activated protein (MAP) kinase phosphatase-4 plays a potential role in insulin resistance. J Biol Chem. 2003;278(32):30187–30192. doi:10.1074/jbc.M302010200
84. Emanuelli B, Eberle D, Suzuki R, Kahn CR. Overexpression of the dual-specificity phosphatase MKP-4/DUSP-9 protects against stress-induced insulin resistance. Proc Natl Acad Sci U S A. 2008;105(9):3545–3550. doi:10.1073/pnas.0712275105
85. Mizuno TM. Fat Mass and Obesity Associated (FTO) gene and hepatic glucose and lipid metabolism. Nutrients. 2018;10(11):1600. doi:10.3390/nu10111600
86. Gerken T, Girard CA, Tung YC, et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science. 2007;318(5855):1469–1472. doi:10.1126/science.1151710
87. Sanchez-Pulido L, Andrade-Navarro MA, The FTO. (fat mass and obesity associated) gene codes for a novel member of the non-heme dioxygenase superfamily. BMC Biochem. 2007;8:23. doi:10.1186/1471-2091-8-23
88. Jia G, Yang CG, Yang S, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582(23–24):3313–3319. doi:10.1016/j.febslet.2008.08.019
89. Han Z, Niu T, Chang J, et al. Crystal structure of the FTO protein reveals basis for its substrate specificity. Nature. 2010;464(7292):1205–1209. doi:10.1038/nature08921
90. Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi:10.1126/science.1141634
91. Dina C, Meyre D, Gallina S, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39(6):724–726. doi:10.1038/ng2048
92. Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi:10.1038/ng.686
93. Hinney A, Nguyen TT, Scherag A, et al. Genome wide association (GWA) study for early onset extreme obesity supports the role of fat mass and obesity associated gene (FTO) variants. PLoS One. 2007;2(12):e1361. doi:10.1371/journal.pone.0001361
94. Yang J, Loos RJ, Powell JE, et al. FTO genotype is associated with phenotypic variability of body mass index. Nature. 2012;490(7419):267–272. doi:10.1038/nature11401
95. Zhang X, Qi Q, Zhang C, et al. FTO genotype and 2-year change in body composition and fat distribution in response to weight-loss diets: the POUNDS LOST Trial. Diabetes. 2012;61(11):3005–3011. doi:10.2337/db11-1799
96. Abdullah A, Peeters A, de Courten M, Stoelwinder J. The magnitude of association between overweight and obesity and the risk of diabetes: a meta-analysis of prospective cohort studies. Diabetes Res Clin Pract. 2010;89(3):309–319. doi:10.1016/j.diabres.2010.04.012
97. Tanaka S, Honda M, Wu B, Kazumi T. Clinical features of normal weight Japanese patients with type 2 diabetes who had formerly been obese. J Atheroscler Thromb. 2011;18(2):115–121. doi:10.5551/jat.5926
98. Park JY, Lee KU, Kim CH, et al. Past and current obesity in Koreans with non-insulin-dependent diabetes mellitus. Diabetes Res Clin Pract. 1997;35(1):49–56. doi:10.1016/S0168-8227(96)01363-0
99. Ragvin A, Moro E, Fredman D, et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 2010;107(2):775–780. doi:10.1073/pnas.0911591107
100. Ihara A, Yamagata K, Nammo T, et al. Functional characterization of the HNF4alpha isoform (HNF4alpha8) expressed in pancreatic beta-cells. Biochem Biophys Res Commun. 2005;329(3):984–990. doi:10.1016/j.bbrc.2005.02.072
101. Nammo T, Yamagata K, Hamaoka R, et al. Expression profile of MODY3/HNF-1alpha protein in the developing mouse pancreas. Diabetologia. 2002;45(8):1142–1153. doi:10.1007/s00125-002-0892-8
102. Marcil V, Delvin E, Sane AT, Tremblay A, Levy E. Oxidative stress influences cholesterol efflux in THP-1 macrophages: role of ATP-binding cassette A1 and nuclear factors. Cardiovasc Res. 2006;72(3):473–482. doi:10.1016/j.cardiores.2006.08.024
103. Parviz F, Matullo C, Garrison WD, et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet. 2003;34(3):292–296. doi:10.1038/ng1175
104. Boj SF, Parrizas M, Maestro MA, Ferrer J. A transcription factor regulatory circuit in differentiated pancreatic cells. Proc Natl Acad Sci U S A. 2001;98(25):14481–14486. doi:10.1073/pnas.241349398
105. Briancon N, Weiss MC. In vivo role of the HNF4alpha AF-1 activation domain revealed by exon swapping. EMBO J. 2006;25(6):1253–1262. doi:10.1038/sj.emboj.7601021
106. Nakhei H, Lingott A, Lemm I, Ryffel GU. An alternative splice variant of the tissue specific transcription factor HNF4alpha predominates in undifferentiated murine cell types. Nucleic Acids Res. 1998;26(2):497–504. doi:10.1093/nar/26.2.497
107. Thomas H, Jaschkowitz K, Bulman M, et al. A distant upstream promoter of the HNF-4alpha gene connects the transcription factors involved in maturity-onset diabetes of the young. Hum Mol Genet. 2001;10(19):2089–2097. doi:10.1093/hmg/10.19.2089
108. Eeckhoute J, Moerman E, Bouckenooghe T, et al. Hepatocyte nuclear factor 4 alpha isoforms originated from the P1 promoter are expressed in human pancreatic beta-cells and exhibit stronger transcriptional potentials than P2 promoter-driven isoforms. Endocrinology. 2003;144(5):1686–1694. doi:10.1210/en.2002-0024
109. Hansen SK, Parrizas M, Jensen ML, et al. Genetic evidence that HNF-1alpha-dependent transcriptional control of HNF-4alpha is essential for human pancreatic beta cell function. J Clin Invest. 2002;110(6):827–833. doi:10.1172/JCI0215085
110. Gardner DS, Tai ES. Clinical features and treatment of maturity onset diabetes of the young (MODY). Diabetes Metab Syndr Obes. 2012;5:101–108. doi:10.2147/DMSO
111. Yamagata K, Furuta H, Oda N, et al. Mutations in the hepatocyte nuclear factor-4alpha gene in maturity-onset diabetes of the young (MODY1). Nature. 1996;384(6608):458–460. doi:10.1038/384458a0
112. Bellanne-Chantelot C, Carette C, Riveline JP, et al. The type and the position of HNF1A mutation modulate age at diagnosis of diabetes in patients with maturity-onset diabetes of the young (MODY)-3. Diabetes. 2008;57(2):503–508. doi:10.2337/db07-0859
113. Cho YS, Chen CH, Hu C, et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44(1):67–72. doi:10.1038/ng.1019
114. Jafar-Mohammadi B, Groves CJ, Gjesing AP, et al. A role for coding functional variants in HNF4A in type 2 diabetes susceptibility. Diabetologia. 2011;54(1):111–119. doi:10.1007/s00125-010-1916-4
115. Bartoov-Shifman R, Hertz R, Wang H, Wollheim CB, Bar-Tana J, Walker MD. Activation of the insulin gene promoter through a direct effect of hepatocyte nuclear factor 4 alpha. J Biol Chem. 2002;277(29):25914–25919. doi:10.1074/jbc.M201582200
116. Rhee J, Inoue Y, Yoon JC, et al. Regulation of hepatic fasting response by PPARgamma coactivator-1alpha (PGC-1): requirement for hepatocyte nuclear factor 4alpha in gluconeogenesis. Proc Natl Acad Sci U S A. 2003;100(7):4012–4017. doi:10.1073/pnas.0730870100
117. Byrne MM, Sturis J, Fajans SS, et al. Altered insulin secretory responses to glucose in subjects with a mutation in the MODY1 gene on chromosome 20. Diabetes. 1995;44(6):699–704. doi:10.2337/diab.44.6.699
118. Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi:10.1126/science.1142358
119. Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316(5829):1336–1341. doi:10.1126/science.1142364
120. Miralles F, Portha B. Early development of beta-cells is impaired in the GK rat model of type 2 diabetes. Diabetes. 2001;50(Suppl 1):S84–S88. doi:10.2337/diabetes.50.2007.S84
121. Louveau I, Gondret F. Regulation of development and metabolism of adipose tissue by growth hormone and the insulin-like growth factor system. Domest Anim Endocrinol. 2004;27(3):241–255. doi:10.1016/j.domaniend.2004.06.004
122. Christiansen J, Kolte AM, Hansen T, Nielsen FC. IGF2 mRNA-binding protein 2: biological function and putative role in type 2 diabetes. J Mol Endocrinol. 2009;43(5):187–195. doi:10.1677/JME-09-0016
123. Groenewoud MJ, Dekker JM, Fritsche A, et al. Variants of CDKAL1 and IGF2BP2 affect first-phase insulin secretion during hyperglycaemic clamps. Diabetologia. 2008;51(9):1659–1663. doi:10.1007/s00125-008-1083-z
124. Duesing K, Fatemifar G, Charpentier G, et al. Evaluation of the association of IGF2BP2 variants with type 2 diabetes in French Caucasians. Diabetes. 2008;57(7):1992–1996. doi:10.2337/db07-1789
125. Huang Q, Yin JY, Dai XP, et al. IGF2BP2 variations influence repaglinide response and risk of type 2 diabetes in Chinese population. Acta Pharmacol Sin. 2010;31(6):709–717. doi:10.1038/aps.2010.47
126. Wu HH, Liu NJ, Yang Z, et al. IGF2BP2 and obesity interaction analysis for type 2 diabetes mellitus in Chinese Han population. Eur J Med Res. 2014;19:40. doi:10.1186/2047-783X-19-40
127. Chistiakov DA, Nikitin AG, Smetanina SA, et al. The rs11705701 G>A polymorphism of IGF2BP2 is associated with IGF2BP2 mRNA and protein levels in the visceral adipose tissue – a link to type 2 diabetes susceptibility. Rev Diabet Stud. 2012;9(2–3):112–122. doi:10.1900/RDS.2012.9.112
128. Ruchat SM, Elks CE, Loos RJ, et al. Evidence of interaction between type 2 diabetes susceptibility genes and dietary fat intake for adiposity and glucose homeostasis-related phenotypes. J Nutrigenet Nutrigenomics. 2009;2(4–5):225–234. doi:10.1159/000259341
129. Chan SH, Lim WK, Michalski ST, et al. Germline hemizygous deletion of CDKN2A-CDKN2B locus in a patient presenting with Li-Fraumeni syndrome. Rev Diabet Stud. 2016;1:16015. doi:10.1038/npjgenmed.2016.15
130. Cunnington MS, Santibanez Koref M, Mayosi BM, Burn J, Chromosome KB. 9p21 SNPs associated with multiple disease phenotypes correlate with ANRIL expression. PLoS Genet. 2010;6(4):e1000899. doi:10.1371/journal.pgen.1000899
131. Qian Y, Lu F, Dong M, et al. Cumulative effect and predictive value of genetic variants associated with type 2 diabetes in Han Chinese: a case-control study. PLoS One. 2015;10(1):e0116537. doi:10.1371/journal.pone.0116537
132. Takeuchi F, Serizawa M, Yamamoto K, et al. Confirmation of multiple risk Loci and genetic impacts by a genome-wide association study of type 2 diabetes in the Japanese population. Diabetes. 2009;58(7):1690–1699. doi:10.2337/db08-1494
133. Cauchi S, Meyre D, Durand E, et al. Post genome-wide association studies of novel genes associated with type 2 diabetes show gene-gene interaction and high predictive value. PLoS One. 2008;3(5):e2031. doi:10.1371/journal.pone.0002031
134. Moritani M, Yamasaki S, Kagami M, et al. Hypoplasia of endocrine and exocrine pancreas in homozygous transgenic TGF-beta1. Mol Cell Endocrinol. 2005;229(1–2):175–184. doi:10.1016/j.mce.2004.08.007
135. Russo L, Lumeng CN. Properties and functions of adipose tissue macrophages in obesity. Immunology. 2018;155(4):407–417. doi:10.1111/imm.2018.155.issue-4
136. Liu Z, Habener JF. Wnt signaling in pancreatic islets. Adv Exp Med Biol. 2010;654:391–419.
137. Maschio DA, Oliveira RB, Santos MR, Carvalho CP, Barbosa-Sampaio HC, Collares-Buzato CB. Activation of the Wnt/beta-catenin pathway in pancreatic beta cells during the compensatory islet hyperplasia in prediabetic mice. Biochem Biophys Res Commun. 2016;478(4):1534–1540. doi:10.1016/j.bbrc.2016.08.146
138. Yao DD, Yang L, Wang Y, et al. Geniposide promotes beta-cell regeneration and survival through regulating beta-catenin/TCF7L2 pathway. Cell Death Dis. 2015;6:e1746. doi:10.1038/cddis.2015.107
139. Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–885. doi:10.1038/nature05616
140. Steinthorsdottir V, Thorleifsson G, Reynisdottir I, et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat Genet. 2007;39(6):770–775. doi:10.1038/ng2043
141. Saxena R, Elbers CC, Guo Y, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90(3):410–425. doi:10.1016/j.ajhg.2011.12.022
142. Acharya S, Al-Elq A, Al-Nafaie A, Muzaheed M, Al-AliA. Type 2 diabetes mellitus susceptibility gene TCF7L2 is strongly associated with hyperglycemia in the Saudi Arabia Population of the eastern province of Saudi Arabia. Eur Rev Med Pharmacol Sci. 2015;19(16):3100–3106.
143. Struewing I, Boyechko T, Barnett C, Beildeck M, Byers SW, Mao CD. The balance of TCF7L2 variants with differential activities in Wnt-signaling is regulated by lithium in a GSK3beta-independent manner. Biochem Biophys Res Commun. 2010;399(2):245–250. doi:10.1016/j.bbrc.2010.07.062
144. Xia Q, Deliard S, Yuan CX, Johnson ME, Grant SF. Characterization of the transcriptional machinery bound across the widely presumed type 2 diabetes causal variant, rs7903146, within TCF7L2. Eur J Hum Genet. 2015;23(1):103–109. doi:10.1038/ejhg.2014.48
145. Shao W, Wang D, Chiang YT, et al. The Wnt signaling pathway effector TCF7L2 controls gut and brain proglucagon gene expression and glucose homeostasis. Diabetes. 2013;62(3):789–800. doi:10.2337/db12-0365
146. Nadkarni P, Chepurny OG, Holz GG. Regulation of glucose homeostasis by GLP-1. Prog Mol Biol Transl Sci. 2014;121:23–65.
147. Cho YM, Kieffer TJ. K-cells and glucose-dependent insulinotropic polypeptide in health and disease. Vitam Horm. 2010;84:111–150.
148. Li R, Ou J, Li L, Yang Y, Zhao J, Wu R. The Wnt signaling pathway effector TCF7L2 mediates olanzapine-induced weight gain and insulin resistance. Front Pharmacol. 2018;9:379. doi:10.3389/fphar.2018.00379
149. Liu H, Fergusson MM, Wu JJ, et al. Wnt signaling regulates hepatic metabolism. Sci Signal. 2011;4(158):ra6. doi:10.1126/scisignal.2001249
150. Ross SE, Hemati N, Longo KA, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289(5481):950–953. doi:10.1126/science.289.5481.950
151. Inagaki N, Gonoi T, Clement J, et al. Reconstitution of IKATP: an inward rectifier subunit plus the sulfonylurea receptor. Science. 1995;270(5239):1166–1170. doi:10.1126/science.270.5239.1166
152. Aguilar-Bryan L, JPt C, Gonzalez G, Kunjilwar K, Babenko A, Bryan J. Toward understanding the assembly and structure of KATP channels. Physiol Rev. 1998;78(1):227–245. doi:10.1152/physrev.1998.78.1.227
153. McTaggart JS, Clark RH, Ashcroft FM. The role of the KATP channel in glucose homeostasis in health and disease: more than meets the islet. J Physiol. 2010;588(Pt 17):3201–3209. doi:10.1113/jphysiol.2010.191767
154. Ashcroft FM. K(ATP) channels and insulin secretion: a key role in health and disease. Biochem Soc Trans. 2006;34(Pt 2):243–246. doi:10.1042/BST0340243
155. Wang DD, Chen X, Yang Y, Liu CX. Association of Kir6.2 gene rs5219 variation with type 2 diabetes: A meta-analysis of 21,464 individuals. Prim Care Diabetes. 2018;12(4):345–353. doi:10.1016/j.pcd.2018.03.004
156. Gloyn AL, Pearson ER, Antcliff JF, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350(18):1838–1849. doi:10.1056/NEJMoa032922
157. Karunakaran U, Park KG. A systematic review of oxidative stress and safety of antioxidants in diabetes: focus on islets and their defense. Diabetes Metab J. 2013;37(2):106–112. doi:10.4093/dmj.2013.37.2.106
158. Chen H, Yu M, Li M, et al. Polymorphic variations in manganese superoxide dismutase (MnSOD), glutathione peroxidase-1 (GPX1), and catalase (CAT) contribute to elevated plasma triglyceride levels in Chinese patients with type 2 diabetes or diabetic cardiovascular disease. Mol Cell Biochem. 2012;363(1–2):85–91. doi:10.1007/s11010-011-1160-3
159. Bid HK, Konwar R, Saxena M, Chaudhari P, Agrawal CG, Banerjee M. Association of glutathione S-transferase (GSTM1, T1 and P1) gene polymorphisms with type 2 diabetes mellitus in north Indian population. J Postgrad Med. 2010;56(3):176–181. doi:10.4103/0022-3859.68633
160. Jha JC, Gray SP, Barit D, et al. Genetic targeting or pharmacologic inhibition of NADPH oxidase nox4 provides renoprotection in long-term diabetic nephropathy. J Am Soc Nephrol. 2014;25(6):1237–1254. doi:10.1681/ASN.2013070810
161. Banerjee M, Vats P, Kushwah AS, Srivastava N. Interaction of antioxidant gene variants and susceptibility to type 2 diabetes mellitus. Br J Biomed Sci. 2019;76(4)1–6.
162. Adachi Y, Yamamoto K, Okada T, Yoshida H, Harada A, Mori K. ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct Funct. 2008;33(1):75–89. doi:10.1247/csf.07044
163. Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12(12):1812–1824. doi:10.1101/gad.12.12.1812
164. Kim I, Xu W, Reed JC. Cell death and endoplasmic reticulum stress: disease relevance and therapeutic opportunities. Nat Rev Drug Discov. 2008;7(12):1013–1030. doi:10.1038/nrd2755
165. Puigserver P, Adelmant G, Wu Z, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi:10.1126/science.286.5443.1368
166. Puigserver P, Wu Z, Park CW, Graves R, Wright M, Spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92(6):829–839. doi:10.1016/S0092-8674(00)81410-5
167. Liang H, Ward WF. PGC-1alpha: a key regulator of energy metabolism. Adv Physiol Educ. 2006;30(4):145–151. doi:10.1152/advan.00052.2006
168. Petersen KF, Befroy D, Dufour S, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300(5622):1140–1142. doi:10.1126/science.1082889
169. Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51(10):2944–2950. doi:10.2337/diabetes.51.10.2944
170. Antonopoulos AS, Margaritis M, Coutinho P, et al. Adiponectin as a link between type 2 diabetes and vascular NADPH oxidase activity in the human arterial wall: the regulatory role of perivascular adipose tissue. Diabetes. 2015;64(6):2207–2219. doi:10.2337/db14-1011
171. Alkhateeb A, Al-Azzam S, Zyadine R, Abuarqoub D. Genetic association of adiponectin with type 2 diabetes in Jordanian Arab population. Gene. 2013;512(1):61–63. doi:10.1016/j.gene.2012.09.095
172. Beltcheva O, Boyadzhieva M, Angelova O, Mitev V, Kaneva R, Atanasova I. The rs266729 single-nucleotide polymorphism in the adiponectin gene shows association with gestational diabetes. Arch Gynecol Obstet. 2014;289(4):743–748. doi:10.1007/s00404-013-3029-z
173. Cox AJ, Lambird JE, An SS, et al. Variants in adiponectin signaling pathway genes show little association with subclinical CVD in the diabetes heart study. Obesity (Silver Spring). 2013;21(9):E456–E462. doi:10.1002/oby.20184
174. Chakraborti CK. Role of adiponectin and some other factors linking type 2 diabetes mellitus and obesity. World J Diabetes. 2015;6(15):1296–1308. doi:10.4239/wjd.v6.i15.1296
175. Koh EH, Park JY, Park HS, et al. Essential role of mitochondrial function in adiponectin synthesis in adipocytes. Diabetes. 2007;56(12):2973–2981. doi:10.2337/db07-0510
176. Chistiakov DA, Potapov VA, Smetanina SA, Bel’chikova LN, Suplotova LA, Nosikov VV. The carriage of risk variants of CDKAL1 impairs beta-cell function in both diabetic and non-diabetic patients and reduces response to non-sulfonylurea and sulfonylurea agonists of the pancreatic KATP channel. Acta Diabetol. 2011;48(3):227–235. doi:10.1007/s00592-011-0299-4
177. Pascoe L, Tura A, Patel SK, et al. Common variants of the novel type 2 diabetes genes CDKAL1 and HHEX/IDE are associated with decreased pancreatic beta-cell function. Diabetes. 2007;56(12):3101–3104. doi:10.2337/db07-0634
178. Liang J, Pei Y, Liu X, et al. The CDKAL1 gene is associated with impaired insulin secretion and glucose-related traits: the Cardiometabolic Risk in Chinese (CRC) study. Clin Endocrinol (Oxf). 2015;83(5):651–655. doi:10.1111/cen.2015.83.issue-5
179. Nfor ON, Wu MF, Lee CT, et al. Body mass index modulates the association between CDKAL1 rs10946398 variant and type 2 diabetes among Taiwanese women. Sci Rep. 2018;8(1):13235.
180. Dubern B, Lubrano-Berthelier C, Mencarelli M, et al. Mutational analysis of the pro-opiomelanocortin gene in French obese children led to the identification of a novel deleterious heterozygous mutation located in the alpha-melanocyte stimulating hormone domain. Pediatr Res. 2008;63(2):211–216. doi:10.1203/PDR.0b013e31815ed62b
181. Lee YS, Challis BG, Thompson DA, et al. A POMC variant implicates beta-melanocyte-stimulating hormone in the control of human energy balance. Cell Metab. 2006;3(2):135–140. doi:10.1016/j.cmet.2006.01.006
182. Mountjoy KG. Functions for pro-opiomelanocortin-derived peptides in obesity and diabetes. Biochem J. 2010;428(3):305–324. doi:10.1042/BJ20091957
183. Parton LE, Ye CP, Coppari R, et al. Glucose sensing by POMC neurons regulates glucose homeostasis and is impaired in obesity. Nature. 2007;449(7159):228–232.
184. Fick LJ, Belsham DD. Nutrient sensing and insulin signaling in neuropeptide-expressing immortalized, hypothalamic neurons: A cellular model of insulin resistance. Cell Cycle. 2010;9(16):3186–3193. doi:10.4161/cc.9.16.12601
185. Butler AA, Kesterson RA, Khong K, et al. A unique metabolic syndrome causes obesity in the melanocortin-3 receptor-deficient mouse. Endocrinology. 2000;141(9):3518–3521. doi:10.1210/endo.141.9.7791
186. Barzilai N, She L, Liu L, et al. Decreased visceral adiposity accounts for leptin effect on hepatic but not peripheral insulin action. Am J Physiol. 1999;277(2):E291–E298. doi:10.1152/ajpendo.1999.277.2.E291
187. Saremi L, Lotfipanah S, Mohammadi M, et al. The Pro12Ala polymorphism in the PPAR-gamma2 gene is not associated with an increased risk of NAFLD in Iranian patients with type 2 diabetes mellitus. Cell Mol Biol Lett. 2019;24:12. doi:10.1186/s11658-019-0138-0
188. Motavallian A, Andalib S, Vaseghi G, Mirmohammad-Sadeghi H, Amini M. Association between PRO12ALA polymorphism of the PPAR-gamma2 gene and type 2 diabetes mellitus in Iranian patients. Indian J Hum Genet. 2013;19(2):239–244. doi:10.4103/0971-6866.116126
189. Gupta AC, Chaudhory AK. Sukriti, et al. Peroxisome proliferators-activated receptor gamma2 Pro12Ala variant is associated with body mass index in non-alcoholic fatty liver disease patients. Hepatol Int. 2010;5(1):575–580. doi:10.1007/s12072-010-9225-z
190. Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi:10.1146/annurev.med.57.121304.131428
191. Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet. 2000;26(1):76–80. doi:10.1038/79216
192. Chan KH, Niu T, Ma Y, et al. Common genetic variants in peroxisome proliferator-activated receptor-gamma (PPARG) and type 2 diabetes risk among Women’s Health Initiative postmenopausal women. J Clin Endocrinol Metab. 2013;98(3):E600–E604. doi:10.1210/jc.2012-3644
193. Phani NM, Vohra M, Rajesh S, et al. Implications of critical PPARgamma2, ADIPOQ and FTO gene polymorphisms in type 2 diabetes and obesity-mediated susceptibility to type 2 diabetes in an Indian population. Mol Genet Genomics. 2016;291(1):193–204. doi:10.1007/s00438-015-1097-4
194. Masugi J, Tamori Y, Mori H, Koike T, Kasuga M. Inhibitory effect of a proline-to-alanine substitution at codon 12 of peroxisome proliferator-activated receptor-gamma 2 on thiazolidinedione-induced adipogenesis. Biochem Biophys Res Commun. 2000;268(1):178–182. doi:10.1006/bbrc.2000.2096
195. Valve R, Sivenius K, Miettinen R, et al. Two polymorphisms in the peroxisome proliferator-activated receptor-gamma gene are associated with severe overweight among obese women. J Clin Endocrinol Metab. 1999;84(10):3708–3712. doi:10.1210/jcem.84.10.6061
196. Chang YC, Liu PH, Yu YH, et al. Validation of type 2 diabetes risk variants identified by genome-wide association studies in Han Chinese population: a replication study and meta-analysis. PLoS One. 2014;9(4):e95045. doi:10.1371/journal.pone.0095045
197. Kang ES, Kim MS, Kim YS, et al. A polymorphism in the zinc transporter gene SLC30A8 confers resistance against posttransplantation diabetes mellitus in renal allograft recipients. Diabetes. 2008;57(4):1043–1047. doi:10.2337/db07-0761
198. Kleiner S, Gomez D, Megra B, et al. Mice harboring the human SLC30A8 R138X loss-of-function mutation have increased insulin secretory capacity. Proc Natl Acad Sci U S A. 2018;115(32):E7642–E7649. doi:10.1073/pnas.1721418115
199. Chimienti F, Devergnas S, Favier A, Seve M. Identification and cloning of a beta-cell-specific zinc transporter, ZnT-8, localized into insulin secretory granules. Diabetes. 2004;53(9):2330–2337. doi:10.2337/diabetes.53.9.2330
200. Ortega RM, Rodriguez-Rodriguez E, Aparicio A, et al. Poor zinc status is associated with increased risk of insulin resistance in Spanish children. Br J Nutr. 2012;107(3):398–404. doi:10.1017/S0007114511003114
201. Lee CC, Haffner SM, Wagenknecht LE, et al. Insulin clearance and the incidence of type 2 diabetes in Hispanics and African Americans: the IRAS Family Study. Diabetes Care. 2013;36(4):901–907. doi:10.2337/dc12-1316
202. Rutter GA, Chimienti F. SLC30A8 mutations in type 2 diabetes. Diabetologia. 2015;58(1):31–36. doi:10.1007/s00125-014-3405-7
203. Scuteri A, Sanna S, Chen WM, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi:10.1371/journal.pgen.0030115
204. Achenbach P, Lampasona V, Landherr U, et al. Autoantibodies to zinc transporter 8 and SLC30A8 genotype stratify type 1 diabetes risk. Diabetologia. 2009;52(9):1881–1888. doi:10.1007/s00125-009-1438-0
205. Cornelis MC, Hu FB. Gene-environment interactions in the development of type 2 diabetes: recent progress and continuing challenges. Annu Rev Nutr. 2012;32:245–259. doi:10.1146/annurev-nutr-071811-150648
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.