Back to Journals » OncoTargets and Therapy » Volume 12
Tumor suppressive functions of LZTFL1 in hepatocellular carcinoma
Received 4 December 2018
Accepted for publication 4 March 2019
Published 10 July 2019 Volume 2019:12 Pages 5537—5544
DOI https://doi.org/10.2147/OTT.S196925
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Shasha Li,* Jingjing Li,* Zujiang Yu
Department of Infectious Disease, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, Henan, People’s Republic of China
*These authors contributed equally to this work
Background: Hepatocellular carcinoma (HCC) is the third leading cause of cancer-related mortality worldwide. The poor survival may be due to tumor recurrence and metastasis. Growing evidence indicates that Leucine Zipper Transcription Factor-like 1 (LZTFL1) plays an important role in tumor progression of several cancers such as lung cancer and gastric cancer.
Methods: Real-time PCR was performed to evaluate LZTFL1 expression level in HCC cell lines and patient specimens. The relationship between LZTFL1 expression and the clinicopathological data of the patients was analyzed. Stable cell lines with overexpressing LZTFL1 were set-up, and the cell proliferation, migration, and invasion abilities were analyzed. The protein expression was measured by Western blotting.
Results: Here, we found LZTFL1 expression was decreased in human HCC specimens and HCC cell lines. Downregulation of LZTFL1 expression was correlated with tumor stage and metastasis. The ectopic overexpression of LZTFL1 inhibited cell proliferation, migration, invasion, and the expression of MMP9. In addition, LZTFL1 suppressed epithelial mesenchymal transition (EMT).
Conclusion: Taken together, our results highlight the tumor suppressive role of LZTFL1 in HCC, suggesting that LZTFL1 may represent a potential therapeutic strategy for treating patients with HCC.
Keywords: hepatocellular carcinoma, HCC, LZTFL1, migration and invasion, epithelial mesenchymal transition, EMT
Introduction
Hepatocellular carcinoma (HCC) is one of the most common types of malignancies and is the third most common cause of cancer-related mortality worldwide,1–3 representing 80–90% of all primary liver cancers. Although advances in HCC diagnosis and treatment, HCC remains largely incurable due to the high rate of recurrence and metastasis. The overall prognosis of HCC patients remains unsatisfactory, with an approximately 12% survival rate at 5 years.4 Therefore, it is critical to investigate the molecular mechanisms underlying the tumorigenesis of HCC in order to develop effective new therapeutic targets and prognostic markers.
Leucine Zipper Transcription Factor-like 1 (LZTFL1), located in the chromosome region 3p21.3, was identified as the tumor suppressor in lung cancer and gastric cancer.5–7 Wei et al showed that LZTFL1 could suppress gastric cancer cell migration and invasion by regulating nuclear translocation of β-catenin, indicating its tumor suppressive role.6 In lung cancer, LZTFL1 was associated with recurrence and poor survival, whereas re-expression of LZTFL1 in lung tumor cells inhibited extravasation/colonization of circulating tumor cells to the lung and inhibited tumor growth in vivo. However, the effect of LZTFL1 in the HCC cells still needs to be elucidated.
In this study, we aimed to investigate the function of LZTFL1 in regulating metastasis and progression in HCC. Our data confirmed that LZTFL1 is downregulated in HCC and associated significantly with stage and metastasis. It was identified that upregulation of LZTFL1 expression suppressed cell proliferation and inhibited cell metastasis, implicating a potential application of LZTFL1 in HCC therapy.
Materials and methods
Clinical specimens and cell lines
Seventy primary HCC specimens and fifty corresponding adjacent non-cancerous counterparts were collected by means of tumor resection at the First Affiliated Hospital of Zhengzhou University (Zhengzhou, China). Tissues were put into liquid nitrogen immediately following surgery and then stored at −80 °C until use. Clinical histopathological data were obtained from patient medical records. This study was conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients involved in the present study. The present study was approved by the Ethics Committee of the First Affiliated Hospital of Zhengzhou University, Zhengzhou, China.
HCC cell lines (HepG2 and Huh7) and an immortalized normal human hepatic cell line (LO2) were purchased from the cell bank of Chinese Academy of Sciences and maintained in Dulbecco’s modified eagle medium supplemented with 10% fetal bovine serum and 1% Penicillin-Streptomycin at 37 °C in a 5% CO2 incubator.
RNA isolation and quantitative real-time polymerase chain reaction
Total RNA was extracted from cell lines or clinical specimens using TRIzol (Invitrogen, USA) and reversely transcribed using PrimeScript™ RT Reagent Kit (TaKaRa, Japan) according to the manufacturer’s protocol. Quantitative real-time PCR was subsequently performed using the Brilliant II SYBR Green QPCR Master Mix (Agilient, USA). GAPDH was used as an internal control as previously reported.8 The primer sequences used were shown in Table 1. The relative expression levels were determined by 2−ΔΔCt method.
 |
Table 1 Primer sequences are shown for all genes tested |
Cell transfection and cell sorting
The LZTFL1 ORF sequence was amplified by PCR using specific primers and cloned into the lentiviral expression vector pWPXL (Addgene) to develop a pWPXL-LZTFL1 recombinant plasmid. Virus packaging was performed by the co-transfection of pWPXL-LZTFL1, packaging plasmid pSPAX2 (Addgene) and envelope plasmid pMD2.G (Addgene) using Lipofectamine 3000 (Invitrogen, USA) in HEK 293T cells. Viruses were harvested 48 hrs after transfection, and viral titers were determined. HepG2 and Huh7 were infected with either recombinant lentivirus constitutively expressing LZTFL1, or control empty vector. After 24 hrs of transfection, the transfectants with control empty vector and LZTFL1 expression vector were sorted by MoFlo XDP (Beckman, USA) based on the expression of green fluorescent protein (GFP).
Cell proliferation assay
CCK-8 assay (Beyotime, China) was used according to the manufacturer’s instructions. Cells were seeded in 96-well plates at 2,000 cells/well in quintuplicate. After 24 h, 10 ml of CCK-8 was added into each well. Subsequently, absorbance at 450 nm was measured with a microplate reader (Bio-Rad, USA). For soft agar assays, 500 cells in growth medium mixed with 0.35% soft agar were plated in triplicate onto a 12-well plate with a 0.5% semi-solid agar basal layer. Fresh medium was added every 7 days. After 2 weeks incubation at 37 °C, colonies in the soft agar were photographed and scored under an inverted microscope.
Cell migration and invasion assays
The cell migration assay was assessed using Transwell chambers with a pore size of 8 μm (Corning Incorporated, USA). 2×104 cells in 200 μl serum-free DMEM were seeded in the upper chamber and 600 μl medium supplemented with 20% FBS was added to the lower chamber. For the matrigel invasion assay, the Transwell chamber was coated with matrigel (BD Biosciences, USA) according to manufacturer’s instructions, and followed the same protocol as for migration assay. After incubated at 37 °C for 24 h, cells were fixed and stained with crystal violet (0.5% in methanol) for 15 min. The migrated and invaded cells were then counted in five randomly selected fields with an inverted microscope. The experiment was repeated three times independently.
Western blot
Cells were washed with PBS and lysed in ice-cold RIPA lysis buffer with protease inhibitor cocktail (Beyotime, China) on ice for 30 mins. Lysates were separated by SDS-PAGE electrophoresis. The proteins in gel were transferred onto polyvinylidene difluoride membranes (Millipore, USA). The membranes were blocked and incubated with primary antibody against LZTFL1 (1:1,000, Abcam, USA), MMP9(1:1,000, Abcam, USA), Vimentin (1:1,000, Abcam, USA), E-cadherin (1:1,000, Abcam, USA) and β-actin (1:5,000, Abcam, USA) in 4 °C overnight and then incubated with goat anti-rabbit IgG-HRP secondary antibody (1:5,000, Abcam, USA) for 1 hr at room temperature. Protein expression as assessed using enhanced chemiluminescent substrate (Pierce, USA) and exposure to chemiluminescent film according to manufacturer’s instructions.
Statistical analysis
Data are expressed as means ± SD. Statistical analyses were made with SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). All experiments were repeated at least three times. A two-tailed Student’s t-test or ANOVA was used to compare differences between groups. The difference was considered as statistically significant when the P-value is less than 0.05.
Results
LZTFL1 was downregulated in HCC and associated significantly with stage and metastasis
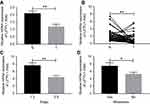
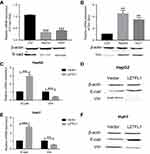
To investigate the expression of LZTFL1 in HCC, 70 HCC specimens and 50 adjacent non-cancerous tissues were analyzed by real-time PCR. The results showed that LZTFL1 expression in HCC tissues was significantly downregulated compared to the adjacent non-cancerous tissues (Figure 1A). Further, 50 paired HCC tissues and corresponding adjacent non-cancerous tissues were analyzed, showing that there was significant downregulation of LZTFL1 mRNA in HCC tissues (Figure 1B). Moreover, the expression of LZTFL1 was significantly correlated with tumor stage (Figure 1C) and metastasis (Figure 1D) but not other variables such as age, gender and differentiation.
LZTFL1 inhibited tumor cell growth in vitro
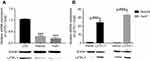
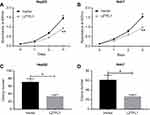
LZTFL1 expression was downregulated in HCC cell lines compared to the immortalized human hepatic cell line (LO2) at mRNA and protein level (Figure 2A). In order to determine whether LZTFL1 plays a direct role in tumorigenesis, we generated stable cell lines that overexpress either control green fluorescent protein or LZTFL1 with GFP in HepG2 and Huh7 cells using a lentiviral transduction system. The expression of LZTFL1 was confirmed by real time PCR and western blot (Figure 2B). We investigated the effect of LZTFL1 expression on the growth of HCC cells under adherent conditions in monolayer cultures using CCK-8 assay and under anchorage-independent conditions in soft agar assays. Upregulation of LZTFL1 in HepG2 (Figure 3A) and Huh7 (Figure 3B) cells significantly inhibited cell proliferation. We then assessed the effect of LZTFL1 on tumor cell growth under anchorage-independent conditions in soft agar assays. The LZTFL1 up-expressing cells showed dramatically reduced the number of colonies upon control cells (Figure 3C and D).
LZTFL1 inhibited cell migration, invasion and the expression of MMP9
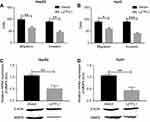
Metastasis is a central problem during cancer treatment. Our result has indicated that the LZTFL1 downregulation was significantly associated with metastasis, as cancer cell migration and invasion are the two major characteristics in the process of metastasis, therefore we aimed to assess whether upregulation of LZTFL1 could affect tumor migration and invasion ability. As shown in Figure 4A and B , the migration and invasiveness of HCC cells were dramatically reduced in LZTFL1 over-expressing cells compared to control cells. MMPs are a family of proteolytic enzymes involved in many phases of cancer progression, including angiogenesis, invasion, and metastasis.9 We found that overexpression of LZTFL1 in HCC cells reduced the expression of MMP9, the major member of MMPs associated with tumor metastasis, both at mRNA and protein levels compared to controls (Figure 4C and D ).
LZTFL1 suppressed the expression of molecular markers associated with EMT
Epithelial mesenchymal transition (EMT) converts polarized, immotile epithelial cells to motile invasive mesenchymal cells.10,11 EMT has been proposed to be a potential mechanism for cancer metastasis.12 The expression of molecular markers associated with EMT was detected in the immortalized human hepatic cell line (LO2) and HCC cell lines (HepG2 and Huh7). As shown in Figure 5A and B, LO2 cells exhibited an increased expression of epithelial cell marker E-cadherin, and a decreased expression of the mesenchymal marker Vimentin. On the contrary, HepG2 and Huh7 cells showed an increased expression of Vimentin, and a decreased expression of E-cadherin. To determine whether LZTFL1 is necessary for HCC cell EMT, we re-expressed of LZTFL1 in HCC cell lines. LZTFL1 overexpression decreased the expression of Vimentin and significantly increased the expression of E-cadherin in HepG2 (Figure 5C and D) and Huh7 cells (Figure 5E and F) both at mRNA and protein levels.
Discussion
HCC is a lethal disease with limited therapeutic options and a particularly poor prognosis. Although significant achievement has been made in the identification of diagnostic and prognostic biomarkers of HCC, our knowledge of the molecular mechanisms underlying HCC development and metastasis is limited. In this study, we revealed that LZTFL1 was downregulated in HCC specimens and that its expression was strongly associated with tumor stage and metastasis. These findings suggest that LZTFL1 has a tumor suppressive function in HCC.
Emerging evidence has demonstrated that decreased LZTFL1 expression results in cancer development.5–7 Here, we found that LZTFL1 is a potential prognostic marker for HCC, because it was downregulated in HCC as compared to adjacent noncancerous tissues and was more highly expressed in non-metastatic than in metastatic tumors. LZTFL1 expression was significantly correlated with tumor stage and metastatic status in HCC. The results from the in vitro assays confirmed that LZTFL1 overexpression inhibited proliferation, migration and invasion of HCC cells, and the expression of MMP9, the major member of MMPs associated with tumor metastasis.13
Metastasis continues to be a lethal hallmark of cancer, with most patients dying as a result of the dissemination of the disease to other organs rather than as a consequence of the primary tumor.14,15 Epithelial mesenchymal transition (EMT) is essential for tumor metastasis and involves a cellular reprogramming process in which epithelial cells dramatically alter their shape, exhibit increased motility and acquire a mesenchymal phenotype.16–18 Loss of expression of epithelial cell markers such as E-cadherin and over expression of mesenchymal cell markers such as Vimentin are a major characteristic of EMT and highly invasive metastatic cancers.19–23 We found that immortalized human hepatic cell line LO2 cells exhibited an increased expression of epithelial cell marker E-cadherin, and a decreased expression of the mesenchymal marker Vimentin. On the contrary, HepG2 and Huh7 cells showed an increased expression of Vimentin, and a decreased expression of E-cadherin. Functional assay showed that re-expression of LZTFL1 could upregulate the expression of the epithelial cell marker E-cadherin. Furthermore, the mesenchymal cell marker Vimentin was downregulated in LZTFL1-overexpressing HCC cells. These data suggest that LZTFL1 may inhibit HCC metastasis by inhibiting the EMT.
In summary, we identified that LZTFL1 is a potential prognostic marker for HCC and LZTFL1 inhibits cell proliferation, migration, invasion and the expression of EMT markers, suggesting its tumor suppressive role in HCC. Therefore, this study provides new insight into the mechanism involved in HCC progression and suggests that LZTFL1 may act as a novel biomarker and promising therapeutic target for HCC.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491.e1.
3. Dang S, Zhou J, Wang Z, Wang K, Dai S, He S. MiR-299-3p functions as a tumor suppressor via targeting Sirtuin 5 in hepatocellular carcinoma. Biomed Pharmacother. 2018;106:966–975. doi:10.1016/j.biopha.2018.06.042
4. Yang Y, Liu Q, Li Z, et al. GP73 promotes epithelial-mesenchymal transition and invasion partly by activating TGF-β1/Smad2 signaling in hepatocellular carcinoma. Carcinogenesis. 2018;39(7):900–910. doi:10.1093/carcin/bgy010
5. Wei Q, Chen Z-H, Wang L, et al. LZTFL1 suppresses lung tumorigenesis by maintaining differentiation of lung epithelial cells. Oncogene. 2016;35(20):2655–2663. doi:10.1038/onc.2015.328
6. Wei Q, Zhou W, Wang W, et al. Tumor-suppressive functions of leucine zipper transcription factor-like 1. Cancer Res. 2010;70(7):2942–2950. doi:10.1158/0008-5472.CAN-09-3826
7. Wang L, Guo J, Wang Q, et al. LZTFL1 suppresses gastric cancer cell migration and invasion through regulating nuclear translocation of β-catenin. J Cancer Res Clin Oncol. 2014;140(12):1997–2008. doi:10.1007/s00432-014-1753-9
8. Wang R, Yang L, Zhang C, et al. Th17 cell-derived IL-17A promoted tumor progression via STAT3/NF-kappaB/Notch1 signaling in non-small cell lung cancer. Oncoimmunology. 2018;7(11):e1461303. doi:10.1080/2162402X.2018.1490854
9. Yadav L, Puri N, Rastogi V, Satpute P, Ahmad R, Kaur G. Matrix metalloproteinases and cancer - roles in threat and therapy. Asian Pac J Cancer Prev. 2014;15(3):1085–1091.
10. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20(2):69–84.
11. Simeone P, Trerotola M, Franck J, et al. The multiverse nature of epithelial to mesenchymal transition. Semin Cancer Biol. 2018. doi:10.1016/j.semcancer.2018.11.004
12. Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer. 2018;18(2):128–134. doi:10.1038/nrc.2017.118
13. Huang H. Matrix metalloproteinase-9 (MMP-9) as a cancer biomarker and MMP-9 biosensors: recent advances. Sensors (Basel). 2018;18(10):3249. doi:10.3390/s18103249
14. Massague J, Batlle E, Gomis RR. Understanding the molecular mechanisms driving metastasis. Mol Oncol. 2017;11(1):3–4. doi:10.1002/1878-0261.12024
15. Celia-Terrassa T, Kang Y. Metastatic niche functions and therapeutic opportunities. Nat Cell Biol. 2018;20(8):868–877. doi:10.1038/s41556-018-0145-9
16. Campbell K. Contribution of epithelial-mesenchymal transitions to organogenesis and cancer metastasis. Curr Opin Cell Biol. 2018;55:30–35. doi:10.1016/j.ceb.2018.06.008
17. Pang A, Carbini M, Moreira AL, Maki RG. Carcinosarcomas and related cancers: tumors caught in the act of epithelial-mesenchymal transition. J Clin Oncol. 2018;36(2):210–216. doi:10.1200/JCO.2017.74.9523
18. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi:10.1038/nrm3758
19. Murai T, Yamada S, Fuchs BC, et al. Epithelial-to-mesenchymal transition predicts prognosis in clinical gastric cancer. J Surg Oncol. 2014;109(7):684–689. doi:10.1002/jso.23564
20. Petrova YI, Schecterson L, Gumbiner BM. Roles for E-cadherin cell surface regulation in cancer. Mol Biol Cell. 2016;27(21):3233–3244. doi:10.1091/mbc.E16-01-0058
21. Giannelli G, Koudelkova P, Dituri F, Mikulits W. Role of epithelial to mesenchymal transition in hepatocellular carcinoma. J Hepatol. 2016;65(4):798–808. doi:10.1016/j.jhep.2016.05.007
22. Messica Y, Laser-Azogui A, Volberg T, et al. The role of vimentin in regulating cell invasive migration in dense cultures of breast carcinoma cells. Nano Lett. 2017;17(11):6941–6948. doi:10.1021/acs.nanolett.7b03358
23. Meng J, Chen S, Han J-X, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–4162. doi:10.1158/0008-5472.CAN-17-3009
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.