Back to Journals » OncoTargets and Therapy » Volume 11
TSPAN7 promotes the migration and proliferation of lung cancer cells via epithelial-to-mesenchymal transition
Authors Wang X, Lin M, Zhao J, Zhu S, Xu M, Zhou X
Received 12 March 2018
Accepted for publication 8 August 2018
Published 13 December 2018 Volume 2018:11 Pages 8815—8822
DOI https://doi.org/10.2147/OTT.S167902
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Xianguo Wang,1,* Min Lin,2,* Jinping Zhao,1 Shaoping Zhu,1 Ming Xu,1 Xuefeng Zhou1
1Thoracic and Cardiovascular Surgery, Zhongnan Hospital of Wuhan University, Wuchang District, Wuhan, Hubei 430071, China; 2Tongji Medical College, Huazhong University of Science and Technology, Wuhan, Hubei 430030, China
*These authors contributed equally to this work
Purpose: To explore the effects and mechanisms of tetraspanin TSPAN7 on the progression of non-small-cell lung cancer (NSCLC) cells.
Patients and methods: All 125 lung cancer specimens and 60 metastatic tissues were obtained from patients diagnosed with NSCLC, and we used immunohistochemistry to detect the expression of TSPAN7 in NSCLC tissues and adjacent normal tissues. Cell proliferation and invasion ability were determined by MTT, colony formation, and cell migration. The relative protein expression level was analyzed by Western blot analysis.
Results: Our clinical data showed that among 125 patients with lung cancer, TSPAN7 was associated with lymph node status, differentiation, tumor size, and poor prognosis. TSPAN7 knockout inhibited cell proliferation and migration. In addition, TSPAN7 increased the expression of N-cadherin in NSCLC cells by reducing the expression of E-cadherin and vimentin and promoting the cell epithelial–mesenchymal transition (EMT) process. Xenograft transplantation model confirmed the role of TSPAN7 in NSCLC metastasis.
Conclusion: TSPAN7-mediated EMT is the key to NSCLC migration. TSPAN7 is a potential target for NSCLC therapy.
Keywords: TSPAN7, non-small-cell lung cancer, cell invasion, colony formation
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. Non-small-cell lung cancer (NSCLC) is the main form of lung cancer and the leading cause of cancer death worldwide, including adenocarcinoma and squamous cell carcinoma.1 Non-small cell lung cancer (NSCLC) patients usually relapse and metastasize after surgery, radiotherapy, and/or chemotherapy, resulting in a 5-year overall survival rate of less than 18%.2 Despite the discovery and improvement of this standard, it is often associated with advanced NSCLC with poor prognosis.3 A persistent problem in NSCLC is the invasion and metastasis of cancer cells, which is the main cause of death.4 Therefore, a detailed understanding of the molecular and cellular mechanisms of metastasis activation is the key to identify new targets for tumor metastasis.
The TM4SF2 gene on XP114 encodes tetraspanin 7 (TSPAN7), which is a member of the tetraspanin protein superfamily of the conserved membrane protein, which is dynamically associated with a number of partner proteins in tetraspanin-enriched microdomains.5 TSPAN7 is widely expressed in nonhematopoietic cells and the strongest expression in the brain.6 TSPAN7-mediated signal transduction plays a role in cell development, activation, growth, and exercise regulation.7 TSPAN7 affects the growth of lung cancer cells in target organ metastasis.8 TSPAN7 might retard the growth of metastases for its relative CD63.9 CD63 is thought to participate in the lysosomal movement and invasive.10 Therefore, we hypothesized that TSPAN7 could affect proliferation and invasion of lung cancer cells. Epithelial–mesenchymal transition (EMT) has been proved to play a more important role in tumor biology in recent years, including inhibition of apoptosis.11 However, there is no evidence that TSPAN7 plays a direct role in the proliferation and invasion of lung cancer cells or its mechanism. The aim of this study was to investigate the mechanism of TSPAN7 on invasion of lung cancer cells and its effect on EMT.
Materials and methods
Patients and specimens
All 125 lung cancer specimens and 60 metastasis tissues were obtained from patients diagnosed with NSCLC and who underwent resection between 2011 and 2016 in Zhongnan Hospital. The study was approved by the institutional review committee of Wuhan University, and all patients signed informed consent. Before surgery, the patients in this study did not receive any treatment, such as radiotherapy or chemotherapy. The patient’s follow-up data are described in each patient’s medical record. The clinical pathological factors are generalized in Table 1. Tumor status, histology, and differentiation were assessed and diagnosed independently by two authors (XG and ML), according to the WHO guidelines of classification.
  | Table 1 Association between TSPAN7 expression and clinicopathologic features of NSCLC patients |
Cell lines and cell culture
Human NSCLC cell lines (H460, A549, H1299, H292) and human normal lung cell line (BEAS-2B) were purchased from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences, Shanghai Institute of Cell Biology. Cells were cultured in RPMI-1640 medium with 10% FBS, 1% penicillin (100 U/mL), and streptomycin (100 μg/mL) (Gibco, Thermo Fisher Scientific, Waltham, MA, USA) at 37°C in a 5% CO2 humid atmosphere.
Plasmids and Transfection
TSPAN7 and empty vector were purchased from Genechem (GENECHEM, Shanghai, China). TSPAN7-shRNA and negative controls were bought from Genechem (GENECHEM). Cells were transfected with plasmids using lipofectamine 2000 reagent (Gibco) and siRNA using X-tremeGENE™ siRNA Transfection Reagent (Sigma-Aldrich Co., St Louis, MO, USA) according to respective manufacturer’s instructions. The transfected efficiency was assessed 48 hours later on protein and mRNA level. All the following experiments after transfection/inference were performed at least in triplicate.
Immunohistochemistry
The cells were fixed with 10% neutral formaldehyde and embedded in paraffin sections. Histological staining of cancer tissues was performed by ABC immunostaining (MaiXin, Fuzhou, China). Then, two independent researchers randomly and blindly evaluated the staining. The criteria for the results: randomly select per slide five views at ×400 magnification with at least 100 cells of each microscopic field.
Western blotting analysis
The cells were lysed using the mammalian protein extraction reagent, RIPA (Beyotime, Shanghai, China). Equivalent amounts (40 μg) of cell protein lysates were electrophoresed on 10% SDS-polyacrylamide gel, transferred to a PVDF membrane. Then the membrane was incubated with primary antibodies with TSPAN7 (1:1,000, Abcam, Massachusetts, USA), N-cadherin (1:2,000, CST, Massachusetts, USA), E-cadherin (1:2,000, CST), vimentin (1:2,000, CST), and tubulin (1:2,000, CST) overnight at 4°C and followed incubation by horseradish peroxidase-labeled secondary antibody. Tubulin was used as a protein-loading control. The immune complexes were tested by chemiluminescence. E-cadherin mouse mAb, N-cadherin rabbit mAb, and β-actin mouse mAb were purchased from Cell Signaling Technology. The cells were lysed in a buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 5 mM EGTA, and 1% NP-40 with a mixture of protease inhibitors before Western blot assay.
Colony formation assays
Cells (500 cells/well) were placed in six-well plates and maintained in media containing 10% FBS. The medium was replaced every 4 days; after 14 days, cells were fixed with methanol and stained with 0.1% crystal violet (Sigma-Aldrich Co.). Visible colonies were then counted. For each treatment group, wells were assessed in triplicate.
Cell migration assays
For the migration assays, at 48 hours post transfection, 5×104 cells in serum-free media were placed into the upper chamber of an insert 8-mm pores and FBS was added to the lower chamber (Millipore, Billerica, MA, USA). Cells that had migrated through the membrane were stained with methanol and 0.1% crystal violet and imaged and counted.
Cell proliferation assay
Cell proliferation rate was determined by using a Cell Counting Kit-8 (CCK-8; CK04 Molecular Technologies, Osaka, Japan). Briefly, cells were seeded into 96-well plates and transfected with overexpression or knockdown plasmid. After transfection for another 24, 48, or 72 hours, 10 μL CCK-8 reagent was added to each well. Cell was incubated for another 3 hours before measuring the absorbance at 450 nm with an enzyme-linked immunosorbent assay plate reader (Bio-Rad). The cell proliferation rate was calculated as the ratio of the OD values of the experimental and control groups. All assays were performed in triplicate.
Nude mice xenograft model
All animals were maintained and used in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Nanjing Medical University. All the experimental procedures were approved by the Nanjing Medical University ethics commission. TSPAN7 overexpression cells and control cells in the logarithmic phase of growth were trypsinized, rinsed with 1× PBS for three times, and then resuspended in 100 μL 1× PBS. Each six nude mice (6 weeks old, male) were inoculated subcutaneously with a clonal population of 5×106 cells in the upper right shoulders subcutaneously. Xenograft tumor sizes were measured by measuring two perpendicular diameters with digital calibers every 7 days after appearance of tumors and calculated according to the formula: 0.5×length×width2. All the mice were sacrificed 1 month after inoculation, and the tumors were removed instantly.
Statistical analysis
All data were expressed as means±SD. SPSS 17.0 software (SPSS Inc., Chicago, IL, USA) was used for the statistical tests. Student’s t-test was used to determine the significance of two group differences. One-way ANOVA was used for multiple comparison. The Pearson correlation coefficient (r) was used for correlation analysis. P<0.05 (two-tailed) was considered to be significant.
Results
High level of TSPAN7 expression associates with clinicopathological factors and poor prognosis in NSCLC patients
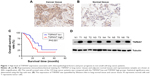
We performed immunohistochemical analysis of 125 lung cancer tissues. TSPAN7 was positive in both adjacent tissues and lung cancer tissues. The expression of TSPAN7 was significantly increased in 80 of the 125 cases (64%) (Figure 1A). In contrast, most normal tissues express low TSPAN7 (Figure 1B). Next, we studied the relationship between TSPAN7 expression level and survival rate. Log rank test showed that TSPAN7 expression was significantly correlated with OS in lung cancer patients (Figure 1C). In order to study the clinical significance of TSPAN7 protein in NSCLC, the relationship between TSPAN7 protein expression and clinicopathological parameters was analyzed. As summarized in Table 1, there was no significant difference in TSPAN7 status with age, sex, and histological differentiation. Tumors with high-regulated TSPAN7 tended to display more aggressive phenotypes, including higher clinical tumor size II–III tumor node metastasis stage, and positive nodal status suggesting an important association between TSPAN7 upregulation and tumor proliferation, invasion, and metastasis. In addition, we found that TSPAN7 was correlated with tumor histology. Meanwhile, we observed that TSPAN7 was upregulated in lung cancer tissues compared with normal cell tissues in the level of protein (Figure 1D).
TSPAN7 is upregulated in human NSCLC cancer cells
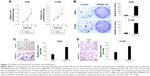
In order to study the expression of TSPAN7 in NSCLC, we first detected the expression of TSPAN7 in four NSCLC cells (H1299, H460, H292, and A549) and normal human lung cell line (BEAS-2B) (Figure 2A). Then, we found that TSPAN7 was upregulated in human NSCLC cancer cells. In order to study lung cancer cells, we transfected plasmids containing TSPAN7 into A549 and H1299 cells, both of which showed low endogenous TSPAN7 expression. Transfection efficiency was confirmed 48 hours after plasmid introduction in Figure 2B and C. We conducted a mixed TSPAN7-specific shRNA transfection to knockout relatively high endogenous expression of TSPAN7 in the H460 and H29 cell lines and then confirm the efficiency of depletion at the protein and RNA levels (Figure 2D).
TSPAN7 overexpression promotes cell proliferation
We observed that overexpression of TSPAN7 promoted cell growth in vitro (Figure 3A and B). Similarly, we found that TSPAN7 overexpression increased the invasiveness of A549 and H1299 cells using Matrigel invasion assays (Figure 3C and D). Overall, these findings suggest that TSPAN7 is an activator of lung cancer progression.
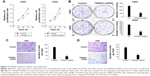
TSPAN7 depletion inhibits cell proliferation
Contrary to the results of TSPAN7 overexpression, TSPAN7 depletion reduced lung cancer growth rate (Figure 4A) and depressed colony formation ability in H460 and H292 cell lines (Figure 4B). Further experiments show that the silencing of TSPAN7 promotes cell migration and invasion in H460 and H29 cell lines (Figure 4C and D).
TSPAN7 affects the expression of EMT markers and promotes NSCLC cells tumorigenesis in vivo
EMT usually leads to pathological migration and invasion in the course of cancer progression. Then, Western blot was used to observe the effect of TSPAN7 on the expression of EMT in A549 and H1299 lung cancer cells. Overexpression of TSPAN7 significantly increases the expression of N-cadherin and vimentin and reduces the expression of E-cadherin in A549 and H1299 cells (Figure 5A). We used H460 and H22 cell experiments to obtain the opposite result of TSPAN7 depletion (Figure 5B). It is suggested that the inhibition of TSPAN7 on invasion and migration of lung cancer cells may be related to EMT. To further clarify the role of TSPAN7 in NSCLC, we will then study the effect of TSPAN7 on tumor growth by using xenograft model. Nude mice were implanted with A549 cells expressing the vector or TSPAN7. The mice were sacrificed in 30 days when the tumor formation became obvious (Figure 5C). Overexpression of TSPAN7 can significantly increase the tumor volume of mice and the growth of NSCLC cells (Figure 5D). The expression of E-cadherin, N-cadherin, and TSPAN7 was detected in the tumor tissue by Western blot. N-cadherin was downregulated and E-cadherin was upregulated in TSPAN7 overexpression in tumor tissues (Figure 5E). These results clearly indicate that TSPAN7 plays a strong role in the tumorigenicity of NSCLC in vivo.
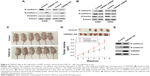  | Figure 5 TSPAN7 affects the expression of EMT markers and promotes NSCLC cells tumorigenesis in vivo. |
Discussion
TSPAN7 promotes the formation of filaments and dendritic spines in cultured hippocampal neurons and is essential for spinal stability and normal synaptic transmission.12 TSPAN7 regulates synaptic transmission and plasticity through an interaction with β1-integrin, PICK1, and AMPA. PICK1 is a mature regulator of AMPAR trafficking and the number of synaptic AMPA receptors.13 TSPAN7 affects actin filament by binding to PI4K13 and/or by β1-integrin.14 The higher the expression of TSPAN7 gene, the lesser the number of TSPAN7-positive blood vessels, the less invasive and metastatic Clear Cell Renal Cell Carcinoma (CCRCC), suggesting that these genes may be the tumor progression and metastasis of tumor growth inhibitors.15 TSPAN7 protein is expected to improve prognosis model based on clinical pathological parameters.15 TSPAN7 may be involved in the development of lung cancer cells.8 Data statistics show that TSPAN7 mRNA has changed in many malignant tumors, especially lung cancer. Our study showed that TSPAN7 protein was upregulated in NSCLC tissues. A direct correlation was observed between upregulation of TSPAN7 and advanced tumor stage, adenocarcinoma, and lymph nodal status. The high expression of TSPAN7 is closely related to the poor prognosis of patients with lung cancer. TSPAN7 deficiency is an independent predictor of OS in NSCLC. At the same time, the level of TSPAN7 in lung cancer cells was significantly higher than that in normal epithelial cells. Overexpression of TSPAN7 promotes growth, proliferation, and invasion of NSCLC cell line. Using shRNA to knock down endogenous TSPAN7, we found that TSPAN7 could promote tumor cell proliferation and invasion.
EMT is one of the most common processes of cell metastasis, which promotes epithelial metastasis and enables epithelial cells to obtain mesenchymal and fibroblast-like properties, including increased mobility and reduced intercellular adhesion.16 EMT is characterized by the loss of epithelial markers, such as E-cadherin, and then gain of mesenchymal markers, such as N-cadherin and vimentin.17 Because of the tumor microenvironment, many EMT pathways are activated by extracellular signals. In our study, we demonstrated that TSPAN7 overexpression in A549 decreased the expression of E-cadherin and increased the expression of N-cadherin and vimentin, which indicated that EMT was promoted by TSPAN7 in lung cancer cells, which indicated that the activator of TSPAN7 mediated the most important biological effects. The role of TSPAN7 in the development tumor is limited.
Conclusion
Our clinical data strongly support the evidence that TSPAN7 is a tumor suppressor of NSCLC. In addition, our study indicates for the first time that TSPAN7 inhibits cell invasion through EMT pathway. Our findings indicate that TSPAN7 plays an important role in cancer suppression in NSCLC and is a new target of anticancer.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Fedewa SA, Miller KD, et al. Cancer statistics for Hispanics/Latinos, 2015. CA Cancer J Clin. 2015;65(6):457–480. | ||
Desantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. | ||
Zhang WC, Chin TM, Yang H, et al. Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nat Commun. 2016;7:11702. | ||
Sun M, Liu XH, Lu KH, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5(6):e1298. | ||
Boucheix C, Rubinstein E. Tetraspanins. Cell Mol Life Sci. 2001;58(9):1189–1205. | ||
Hosokawa Y, Ueyama E, Morikawa Y, Maeda Y, Seto M, Senba E. Molecular cloning of a cDNA encoding mouse A15, a member of the transmembrane 4 superfamily, and its preferential expression in brain neurons. Neurosci Res. 1999;35(4):281–290. | ||
Boucheix C, Duc GH, Jasmin C, Rubinstein E. Tetraspanins and malignancy. Expert Rev Mol Med. 2001;2001:1–17. | ||
Wuttig D, Baier B, Fuessel S, et al. Gene signatures of pulmonary metastases of renal cell carcinoma reflect the disease-free interval and the number of metastases per patient. Int J Cancer. 2009;125(2):474–482. | ||
Pols MS, Klumperman J. Trafficking and function of the tetraspanin CD63. Exp Cell Res. 2009;315(9):1584–1592. | ||
Forte D, Salvestrini V, Corradi G. The tissue inhibitor of metalloproteinases-1 (TIMP-1) promotes survival and migration of acute myeloid leukemia cells through CD63/PI3K/Akt/p21 signaling. Oncotarget. 2016;8(2):2261–2274. | ||
Wu Y, Zhou BP. Snail: more than EMT. Cell Adh Migr. 2010;4(2):199–203. | ||
Bassani S, Passafaro M. TSPAN7: a new player in excitatory synapse maturation and function. Bioarchitecture. 2012;2(3):95–97. | ||
Hanley JG. PICK1: a multi-talented modulator of AMPA receptor trafficking. Pharmacol Ther. 2008;118(1):152–160. | ||
Berditchevski F. Complexes of tetraspanins with integrins: more than meets the eye. J Cell Sci. 2001;114(Pt 23):4143–4151. | ||
Wuttig D, Zastrow S, Füssel S, et al. CD31, EDNRB and TSPAN7 are promising prognostic markers in clear-cell renal cell carcinoma revealed by genome-wide expression analyses of primary tumors and metastases. Int J Cancer. 2012;131(5):E693–E704. | ||
Li H, Ouyang R, Wang Z, et al. MiR-150 promotes cellular metastasis in non-small cell lung cancer by targeting FOXO4. Sci Rep. 2016;6:39001. | ||
Kim KK, Kugler MC, Wolters PJ, et al. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci U S A. 2006;103(35):13180–13185. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.