Back to Journals » OncoTargets and Therapy » Volume 13
TRIM47 Promotes the Development of Glioma by Ubiquitination and Degradation of FOXO1
Authors Wei H, Ding C, Zhuang H, Hu W
Received 28 May 2020
Accepted for publication 8 December 2020
Published 31 December 2020 Volume 2020:13 Pages 13401—13411
DOI https://doi.org/10.2147/OTT.S264459
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr XuYu Yang
Huaming Wei,1 Chonglan Ding,2 Huanxia Zhuang,3 WeiLi Hu4
1Department of Neurology, Jiyang District People’s Hospital of Jinan, Jinan, Shandong 251400, People’s Republic of China; 2Special Inspection Section, Shandong Zaozhuang Traditional Chinese Medicine Hospital, Zaozhuang, Shandong 277000, People’s Republic of China; 3Department of Neurology, Gaotang County People’s Hospital, Gaotang, Shandong 252800, People’s Republic of China; 4Department of Neurology, Lianshui County People’s Hospital, Lianshui, Jiangsu 223400, People’s Republic of China
Correspondence: WeiLi Hu
Department of Neurology, Lianshui County People’s Hospital, No. 6, Hongri Avenue, Lianshui, Jiangsu 223400, People’s Republic of China
Email [email protected]
Objective: To investigate the effect of TRIM47 on glioma cells and further explore its underlying molecular mechanisms.
Methods: Mouse xenograft model was used in this study. The mRNA expression of TRIM47 was detected by qRT-PCR. The cell viability and proliferation activity was detected by MTT assay and colony formation assay. The migration and invasion of glioma cells were determined by Transwell assay. The protein levels of TRIM47, FOXO1, CyclinD1, C-myc, MMP-2 and TIMP-1 were assessed by Western-blotting. The interaction between TRIM47 and FOXO1 was measured by Co-immunoprecipitation (Co-IP) assay.
Results: In glioma tissues and cells, TRIM47 was significantly up-regulated. Silencing the expression of TRIM47 inhibited the cell viability and proliferation of cells A172 and U251, as well as their ability to invade and migrate. Among them, the expression levels of C-myc and CyclinD1 also decreased, and MMP-2 was down-regulated and TIMP-1 was up-regulated. Similarly, in vivo model, tumor volume and weight also decreased after TRIM47 knockout. Further research showed that TRIM47 inhibited FOXO1 expression by ubiquitination and degradation of FOXO1, thereby promoting glioma growth and progression.
Conclusion: In our study, we confirmed functional role of the TRIM47-FOXO1 axis in the progression of gliomas and provided a potential target for glioma treatment.
Keywords: glioma, tripartite motif 47, forkhead box O1, proliferation, migration, invasion
Introduction
Malignant glioma is a primary tumor of the central nervous system, accounting for about 80% of the intracranial malignancies, with the highest morbidity and mortality.1,2 Malignant glioma exhibited increased levels of invasive growth, and is easy to invasion and metastasis.3 At present, the main clinical treatment methods for gliomas are surgical resection plus radiotherapy and chemotherapy. However, because glioma stem cells can resist radiotherapy and chemotherapy, their recurrence rate is still high, and very few patients survive>1 year. In addition, glioma cannot be completely cured, and its prognosis is poor.4 Therefore, it is urgent to study the potential molecular mechanisms and explore new potential diagnostic or therapeutic targets for the treatment of glioma.
A growing body of clinical evidence indicated that ubiquitin-mediated degradation of oncogene products or tumor suppressors may be related to the cause of cancer.5 In the ubiquitin-proteasome system (UPS) component, the E3 ubiquitin ligase, which recognized the most specific substrates, was considered a potential diagnostic and therapeutic target for cancer.6 Tripartite motif (TRIM) family proteins are evolutionarily conserved proteins consisting of circular finger domains with one or two b-box domains and a related coiled spiral domain in the amino-terminal region.7 It is involved in a wide range of cell processes, including cell proliferation,8 differentiation,9 apoptosis,10 cell cycle regulation, carcinogenesis,5 etc. Most TRIM proteins confer ubiquitin E3 ligase activity and promote posttranslational modification. Among them, TRIM47 was first identified as overexpression in astrocytoma. In addition, other studies have shown that the high expression of TRIM47 was closely related to the occurrence of human prostate cancer11 and non-small cell lung cancer.12 However, so far, the pathological and clinical role of TRIM47 in glioma has not been revealed.
Forkhead box O1 (FOXO1) belongs to the FOXO transcription factor family and is located on chromosome 13q14, characterized by a conserved winged helix dNA binding domain.13 In recent years, the broad role of FOXO in physiological processes has been extensively studied, including cell cycle arrest, angiogenesis, apoptosis, stem cell differentiation and stress resistance.14 Previous studies have shown that FOXO1 has an anticancer effect and has been identified as an anticancer gene. In addition, there is increasing evidence that FOXO1 is downregulated in many cancer types, such as Hodgkin’s lymphoma,15 breast cancer,16 and alveolar rhabdomyosarcoma.17 Notingly, although many studies have linked FOXO1 expression to glioma, the exact pattern and role of FOXO1 in glioma remains elusive.
More importantly, no one has studied the complex functions of FOXO1 and TRIM47 in the development and progression of gliomas. Hence, our current study aims to investigate the correlation between TRIM47 and FOXO1 in glioma and to explore its potential mechanisms.
Materials and Methods
Clinical Sample Collection
Seventy-nine glioma patients were selected to obtain paired glioma tissue samples and corresponding adjacent tissue samples. The glioma tissue samples were classified as either low-grade glioma (WHO I–II) or high-grade glioma (WHO III–IV) according to the clinicopathologist’s diagnosis. Each patient signed a written informed consent and the ethical committee of Lianshui County People’s Hospital (LSXRM2019-04).
Cell Culture
Human normal glial cell line (HEB) and five glioma cell lines (U251, Hs683, U87,SHG-44 and A172) were obtained from Mlbio (Shanghai, China). All cells were cultured in RPMI 1640 medium supplemented with 10% FBS in a humidified environment. Subsequent transfection experiments could be performed when cells reached 70% ~ 80% confluences.
Cell Transfection
A172 and U251 with a confluence of 70%~80% were collected and then inoculated into 6-well plates, respectively. They were divided into 5 groups: Mock group (without any treatment), si-NC group (transfected with siRNA NC), si-TRIM47#1 (transfected with siRNA1), si-TRIM47#2 (transfected with siRNA2) and si-TRIM47#3 (transfected with siRNA3), and then follow the instructions of Lipofectamine 2000 liposome transfection kit (Invitrogen, USA). After 48 h transfection, the cells were collected and then for further experiments.
Real-Time PCR Analysis
Total RNA was isolated from glioma tissues and cell lines using TRIzol reagent (Invitrogen, USA). The extracted RNA was reversely transcribed to cDNA by using a Prime Script™ RT Reagent Kit (TaKaRa, Japan). Subsequently, RT-qPCR was performed. The primer sequences were as follows: TRIM47 sense, 5′-GCTTCAGGAGGCTGAGCAGT-3′ and antisense, 5′-TCTGCTACGGCTGCACTCTT-3′; FOXO1 sense, 5′-TACGAGTGGATGGTCAAGAG-3′and antisense, 5′-ATGAACTTGCTGTGTAGGGAC-3′; GAPDH sense, 5′-GCATTGCCCTCAACGACCAC-3′and antisense, 5′- CCACCACCCTGTTGCTGTAG-3′.
Western Blotting
According to the manufacture’s instruction, the proteins were extracted and its concentration was measured. Protein samples were separated by polyacrylamide-SDS gels and electro blotted onto nitrocellulose membranes. After blocking with 5% nonfat dry milk for 2.5 h, the PVDF membrane incubated with primary antibodies: cyclinD1 (1:1000, Sigma Aldrich, SAB4502603), C-myc (1:1000, Thermo Fisher Scientific, 13–2500), MMP-2 (1:1000, Sigma Aldrich, SAB4501891), TIMP-1 (1:1000, Sigma Aldrich, AB770), TRIM47 (1:1000, Sigma Aldrich, SAB2108331), FOXO1 (1:1000, Thermo Fisher Scientific, MA5-14,846). On the following day, the protein samples were incubated with the secondary antibody at room temperature for 45 min, after which blots were visualized by enhanced chemiluminescence method. The intensity of bands was quantified by ImageJ software (Biorad, Richmond, CA).
Co-Immunoprecipitation (Co-IP) Assay
For co-immunoprecipitation assay, IgG or IP antibody, suspended IP Matrix and PBS were incubated 1 hour at 4 °C, following by centrifugation and washing by PBS containing protease inhibitor, phosphatase inhibitor and PMSF for 3 times, and then discard the supernatant. Subsequently, the transfected cells were lysed and transferred to the matrix, and then cells were incubated at 4 °C overnight. The matrix was then centrifuged and washed for 5 times. The SDS-PAGE sample loading buffer was added to the immunoprecipitates, and then was boiled for 10 min at 100 °C. The IP and Input proteins were detected by Western blot.
Cell Viability Assay
Cell viability in each group was assessment by MTT and the transfection time was 0 h. The cells were seeded in a 96-well plate and cultured in a 37°C incubator containing 5% CO2. First, 20 μL of MTT solution (5mg/mL, Sigma) was added to each well, and then continued to incubate in a 5% CO2 incubator at 37°C. After 4h, the culture was terminated and 150 μL of DMSO was added. Gently evenly shake the mixture for 10 min to promote crystallization dissolution. The absorbance values of each hole at 570 nm wavelength were measured on a microplate and data were expressed as absorbance.
Colony Formation Assay
The same quantities of A172 and U251 cells of each group were collected and cultured in 6-well plates. The medium was changed every 2 to 3 days and cultured continuously for 14 days. After washing the cells twice with PBS, the cells were fixed with 4% formaldehyde for 15 min at 37°C. Subsequently, residual formaldehyde was removed, and cell clones were stained with crystal violet for 10–20 min. Cells in each hole were observed under the microscope and the number of clones was counted. Each group≥50 cells were recorded as effective clones, and cameras took photos directly.
Transwell Assay
A172 and U251 cells were trypsinized and re-suspended in serum-free medium to adjust cell density. Subsequently, a total of 1×105 cells were added to the Transwell chamber coated with Matrigel (BD Biosciences, New Jersey, USA), while a medium containing 15% fetal bovine serum was added to the lower chamber. After 24 h in the incubator, the cells were rinsed with PBS, fixed with 4% paraformaldehyde 37 °C for 16 min and stained with 0.1% crystal violet at 37 °C for 25 min. The cell invasion assay was performed in a similar manner, but 20 μL Matrigel (BD Biosciences) was added to the upper chamber. Five fields were randomly selected under an inverted microscope to represent the invasion ability and migration ability of cells in each group.
Mouse Xenograft Model
To establish the xenograft model, 1×106 U251 cells were subcutaneously injected into BALB/c mice (4-week-old). When tumors were measurable, the mice were randomly divided into 3 groups, and PBS, si-NC and si-TRIM47 viruses were injected into the tumor every 2 days for 14d. After 30 days, tumor volume and tumor weight were recorded.
Immunohistochemistry (IHC) Staining
The xenograft tumors were formalin-fixed and paraffin-embedded, sliced into 4µm-thick sections. The xenograft tumors were formalin-fixed and paraffin-embedded, sliced into 4 µm-thick sections. Graded ethanol was used to rehydrate the sections after deparaffinization in xylene at room temperature. After washing with PBS, the sections were placed in 3% hydrogen peroxide for 20 min to inhibit endogenous peroxidase, followed by antigen retrieval by heating for 30 min in a microwave. After being blocked with 10% goat sera, the tissue sections were incubated with primary anti-Ki67 antibody (1:500, Cell Signaling Technology, MA, USA) and at 4°C overnight. After being washed with PBS and incubated with secondary antibody for 30 min at 37°C, 3,′3-Diaminobenzidine tetrahydrochloride (DAB) was applied as a chromogen, and the sections were counterstained with hematoxylin.
Statistical Analysis
The data were presented as the mean ± standard deviation (SD) and analyzed by the Student’s t-test and one-way analysis of variance (ANOVA) using the SPSS version 19.0 software. Survival rate was analyzed by Kaplan-Meier analysis. P<0.05 was considered to be statistically significant.
Results
TRIM47 Was Highly Expressed in Glioma Tissues and Glioma Cell Lines
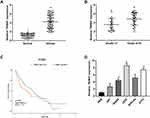
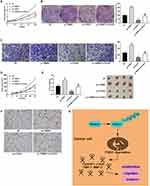
As presented in Figure 1A, the mRNA expression of TRIM47 in glioma tissues were obviously higher than that in the adjacent normal tissue (P<0.05). Notably, compared with grades I–II, the level of TRIM47 was significantly increased in grades III–IV, indicating that the level of TRIM47 expression was closely related to the clinical grade of glioma (Figure 1B; Table 1). Subsequently, we analyzed the overall survival based on the TCGA database, and found that the glioma patients with high expression of TRIM47 had a worse overall survival (Figure 1C). At the same time, we also tested the expression of TRIM47 in different glioma cell lines (U251, Hs683, U87, SHG-44 and A172). Compared to human normal glial cell line (HEB), TRIM47 was significantly up-regulated in the above cell lines, with U251 and A172 being the most pronounced (Figure 1D). Therefore, U251 and A172 were selected for follow-up experiments.
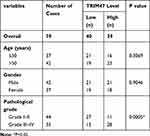 |
Table 1 The Correlation Between TRIM47 mRNA Expression and Clinicopathological Factors |
TRIM47 Promoted the Proliferation of Glioma Cell in vitro and in vivo
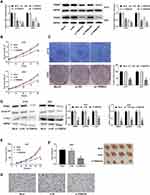
To further investigate the role of TRIM47 on glioma, U251 and A172 cells were then transfected with si-TRIM47 or si-NC. From Figure 2A, we can see that compared with group si-NC, the silencing efficiency of TRIM47 gene in si-TRIM47 groups were very significant, especially in group si-TRIM47#3. Next, we selected group si-TRIM47#3 for MTT assay to evaluate the cell viability of A172 and U251 cells. The MTT results indicated that the above cell viability had been significantly reduced in a time-dependent manner (Figure 2B). Similarly, the colony formation assay also indicated that the low expression of TRIM47 resulted in decreased cell proliferation and colony number, suggesting that si-TRIM47 inhibited the proliferation of A172 and U251 cells (Figure 2C).
C-myc and Cyclin D1 were all proliferation-related proteins, to further detect the effect of TRIM47 in the progression of glioma, Western blotting assay was conducted. As expected, the protein expression of C-myc and Cyclin D1 were decreased in TRIM47 knockdown cells when in comparison to Mock or si-NC groups (Figure 2D). Consistently, down-regulation of TRIM47 sharply reduced the tumor volume (Figure 2E) and tumor weight (Figure 2F). The result of IHC staining showed that there were less positive staining for Ki67 in si-TRIM47 group compared with that in the Mock or si-NC groups (Figure 2G).
TRIM47 Promoted the Migration and Invasion of Glioma Cell
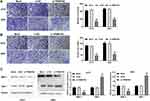
To investigate the role of TRIM47 in migration and invasion of glioma cell, Transwell assay was conducted after transfecting si-NC or si-TRIM47. As shown in Figure 3A and B, si-TRIM47 markedly reduced the number of cell migration and invasion when compared with the Mock or si-NC groups. These results indicate that si-TRIM47 may inhibit migration and invasion of A172 and U251 cells.
MMP-2 and TIMP-1 were all migration-related proteins, and their functions are opposite. Among them, MMP-2 can promote the migration and invasion of tumor cells, while TIMP-1, on the contrary, can inhibit the infiltration and migration of tumor cells. To further clarify the changes of the above proteins in the TRIM47 down-regulated group, we conducted Western blotting. As indicated in Figure 3C, MMP-2 was sharply reduced in si-TRIM47 group, while TIMP-1 protein expression increased significantly in comparison to Mock or si-NC groups.
TRIM47 Interacted with FOXO1 and Enhanced Ubiquitylation and Degradation of FOXO1
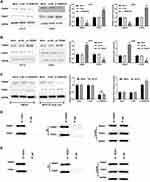
To further understand the potential molecular mechanism of TRIM47 in glioma progression, we first detected the expression of TRIM47 and FOXO1 in glioma cells A172 and U251. As presented in Figure 4A, compared with Mock or si-NC groups, the protein expression of TRIM47 was decreased in A172 and U251 cells, whereas the protein expression of FOXO1 was increased with TRIM47 knockdown (P˂0.05). In addition, up-regulating the expression of TRIM47 remarkably inhibited the protein expression of FOXO1 (Figure 4B). Next, the molecular mechanism by which TRIM47 regulated the expression level of FOXO1 protein was further explored.
Previously, TRIM47 has been reported to have an E3 ubiquitin ligase effect, so we speculated whether TRIM47 might down-regulate FOXO1 expression by promoting ubiquitination and degradation of FOXO1 protein. To prove this, MG132 (proteasome inhibitor, 10 μL/mL) or DMSO was added to treat glioma cells for 2h. The western-blotting results showed that, compared with DMSO group, TRIM47-induced down-regulation of FOXO1 was counteracted by MG132 (Figure 4C, P˂0.05). Furthermore, the Co-IP assay showed that TRIM47 and FOXO1 interacted with each other in A172 and U215 cells (Figure 4D and E).
TRIM47 Promoted Glioma Development Through FOXO1 Ubiquitination
To further investigate the relationship between TRIM47 and FOXO1 in the progression of glioma, MTT assay, Colony formation assay, Transwell assay and Mouse xenograft model were next conducted. As exhibited in Figure 5A, overexpression of TRIM47 significantly increased the cell viability of glioma cells, while overexpression of FOXO1 sharply inhibited the cell viability of glioma cells. It was noted that when pc-TRIM47 and pc-FOXO1 were co-transfected with glioma cells, their cell viability was also significantly reduced, which was still higher than that of pc-FOXO1 group. Consistently, pc-TRIM47 group had the largest number of cell clones, followed by pc-TRIM47+pc-FOXO1group, and pc-FOXO1 group had the least number of clones (Figure 5B). In Figure 5C, we can clearly see that co-transfection pc-TRIM47 and pc-FOXO1 could to some extent offset the migration of glioma cells induced by overexpression TRIM47. In addition, evidence that the TRIM47 and FOXO1 interactions affected tumor progression can be confirmed in mouse xenograft model. Overexpression of TRIM47 increased tumor volume and tumor weight, while pc-FOXO1 has the opposite effect, it would inhibit tumor development. When the two interact with glioma cells, the effects of the two are partially offset (Figure 5D and E). The result of HIC staining showed that TRIM47 overexpression increased the positive staining for Ki67 of tumor tissues, while overexpression of FOXO1 revealed the opposite effect (Figure 5F, P<0.05). FOXO1 overexpression could eliminated the effect of TRIM47 overexpression on the xenograft tumor (Figure 5F, P<0.05).
Discussion
Worldwide, glioma is a life-threatening primary brain cancer. Its uncontrollable growth and expansion are the main reasons for its destruction of surrounding normal brain tissue and damage to nerve function.18 Despite advances in chemotherapy, radiation, and surgery for gliomas over the past few decades, there is currently no effective treatment to improve long-term survival of glioma patients. Therefore, there is an urgent need to elucidate the biological and molecular mechanisms of the development of gliomas, and to develop more effective treatments on this basis.
TRIM47 is an E3 ubiquitin ligase, located at 17q24-25, which is frequently acquired or amplified in many other types of tumors.6 Recent pathological analysis showed that changes in the expression of certain TRIM proteins are closely related to the malignancy of the tumor and the prognosis.19 In our study, we observed that TRIM47 was highly expressed in glioma tissue and cells, and its expression level was significantly correlated with pathological stage. In addition, compared with patients with low TRIM47 expression, the overall survival rate of patients with high TRIM47 expression was significantly reduced. Therefore, we speculate that TRIM47 may play an important role in glioma development. To verify this hypothesis, we then constructed TRIM47 low expression vectors, and evaluated the effect on the proliferation ability and migration ability of glioma cells by silencing TRIM47 expression. As expected, in glioma cells cultured in vitro, silencing the expression of TRIM47 significantly inhibited the proliferation and migration of glioma cells. Consistent with this, tumor volume and tumor weight were also significantly reduced in xenograft nude mouse models after silencing TRIM47 expression. C-myc20 and Cyclin D121 were all proliferation-related proteins, and changes in the expression of these proteins can affect tumorigenesis and progression. MMP-2 and TIMP-1 were both migration-related proteins, but they play the opposite role in tumor progression. It has been reported that MMP-2 can promote the development of a variety of tumors,22,23 and its expression level increased with the increase of malignant potential; while TIMP-1 was to inhibit tumor invasion and metastasis.24 Since the expression change of TRIM47 affected the proliferation, migration and invasion of glioma, does it play a role through the above proteins? With this in mind, we examined changes in the expression levels of the aforementioned proteins. Our results showed that after TRIM47 knockout, the protein levels of C-Myc, Cyclin D1 and MMP-2 significantly decreased, whereas the protein levels of TIMP-1 increased, which was consistent with previous reports. Collectively, the above data suggested that TRIM47 may be an oncogene in human glioma.
TRIM proteins are typically involved in cancer development through ubiquitination and degradation of target proteins.25 However, the potential molecular mechanisms of TRIM47 in glioma cell growth, migration and invasion have been rarely investigated. FOXO1 is a transcription factor that belongs to the FOX family. The gene encoding FOXO1 is located on chromosome 13q14, and methylation, mutation, and allele loss are common in cancer. These characteristics suggest that there are potential tumor-related genes involved in the development of human malignancies in this region.26 A growing body of research reported that FOXO1 as a cancer inhibitor can inhibit the development of different types of cancer, and the inactivation of FOXO1 is related to the poor prognosis of patients.26,27 In addition, FOXO1 protein levels and transcriptional activation are tightly regulated by a variety of post-translational modifications, including phosphorylation, acetylation, ubiquitination, and methylation.28 In the presented study, we concluded that TRIM47 can interact with FOXO1 and degrade FOXO1. First, we found that knocking down TRIM47 increased the expression of FOXO1 in glioma cells, whereas overexpression of TRIM47 decreased the expression of FOXO1. Secondly, treatment with proteasome inhibitor MG132 disrupted TRIM47-induced downer-gulation of FOXO1 in glioma cells. To further explore the role of the relationship between TRIM47 and FOXO1 in glioma progression, pc-TRIM47 and pc-FOXO1were used for subsequent MTT, colony formation assay, Transwell assay experiments. The above data consistently demonstrated that TRIM47 interacted with FOXO1 to degrade FOXO1 to promote the development and progression of glioma.
Conclusion
In our study, TRIM47 was up-regulated in glioma cells. TRIM47 inhibited FOXO1 by ubiquitination and degradation of FOXO1, thereby promoting glioma growth and progression. This indicated that TRIM47 can be considered as a biomarker to guide the diagnosis and treatment of glioma patients, and the measurement of TRIM47 expression may become an effective method for predicting the prognosis of patients.
Statement of Ethics
This study was authorized by the ethical committee of Lianshui County People’s Hospital (LSXRM2019-04) and all patients provided written informed consent.
Ethical and legal approval was obtained prior to the commencement of the study by the ethical committee of Lianshui County People’s Hospital (LSXRM2019-04). All experiments were performed following the ethical committee of Lianshui County People’s Hospital and national guidelines and regulations. All patients provided written informed consent.
Funding
There is no funding to report.
Disclosure
The authors indicate no potential conflicts of interest.
References
1. Shirai K, Chakravarti A. Towards personalized therapy for patients with glioblastoma. Expert Rev Anticancer Ther. 2011;11(12):1935–1944. doi:10.1586/era.11.103
2. Zhang ZS, Wang J, Shen YB, et al. Dihydroartemisinin increases temozolomide efficacy in glioma cells by inducing autophagy. Oncol Lett. 2015;10(1):379–383. doi:10.3892/ol.2015.3183
3. Diao B, Liu Y, Zhang Y, Yu J, Xie J, Xu G-Z. IQGAP1‑siRNA inhibits proliferation and metastasis of U251 and U373 glioma cell lines. Mol Med Rep. 2017;15(4):2074–2082. doi:10.3892/mmr.2017.6257
4. Patil CG, Eboli P, Hu J. Management of multifocal and multicentric gliomas. Neurosurg Clin N Am. 2012;23(2):343–350, x. doi:10.1016/j.nec.2012.01.012
5. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. doi:10.1038/nrc3139
6. Liang Q, Tang C, Tang M, Zhang Q, Gao Y, Ge Z. TRIM47 is up-regulated in colorectal cancer, promoting ubiquitination and degradation of SMAD4. J Exp Clin Cancer Res. 2019;38(1):159. doi:10.1186/s13046-019-1143-x
7. Reymond A, Meroni G, Fantozzi A, et al. The tripartite motif family identifies cell compartments. EMBO J. 2001;20(9):2140–2151. doi:10.1093/emboj/20.9.2140
8. Chen L, Chen DT, Kurtyka C, et al. Tripartite motif containing 28 (Trim28) can regulate cell proliferation by bridging HDAC1/E2F interactions. J Biol Chem. 2012;287(48):40106–40118. doi:10.1074/jbc.M112.380865
9. Sato T, Okumura F, Ariga T, Hatakeyama S. TRIM6 interacts with Myc and maintains the pluripotency of mouse embryonic stem cells. J Cell Sci. 2012;125(Pt 6):1544–1555. doi:10.1242/jcs.095273
10. Zaman MM, Nomura T, Takagi T, et al. Ubiquitination-deubiquitination by the TRIM27-USP7 complex regulates tumor necrosis factor alpha-induced apoptosis. Mol Cell Biol. 2013;33(24):4971–4984. doi:10.1128/MCB.00465-13
11. Fujimura T, Inoue S, Urano T, et al. Increased expression of tripartite motif (TRIM) 47 is a negative prognostic predictor in human prostate cancer. Clin Genitourin Cancer. 2016;14(4):298–303. doi:10.1016/j.clgc.2016.01.011
12. Han Y, Tian H, Chen P, Lin Q. TRIM47 overexpression is a poor prognostic factor and contributes to carcinogenesis in non-small cell lung carcinoma. Oncotarget. 2017;8(14):22730–22740. doi:10.18632/oncotarget.15188
13. Ying H, Kang Y, Zhang H, et al. MiR-127 modulates macrophage polarization and promotes lung inflammation and injury by activating the JNK pathway. J Immunol. 2015;194(3):1239–1251. doi:10.4049/jimmunol.1402088
14. Srivastava RK, Unterman TG, Shankar S. FOXO transcription factors and VEGF neutralizing antibody enhance antiangiogenic effects of resveratrol. Mol Cell Biochem. 2010;337(1–2):201–212. doi:10.1007/s11010-009-0300-5
15. Xie L, Ushmorov A, Leithäuser F, et al. FOXO1 is a tumor suppressor in classical Hodgkin lymphoma. Blood. 2012;119(15):3503–3511. doi:10.1182/blood-2011-09-381905
16. Wu Y, Elshimali Y, Sarkissyan M, Mohamed H, Clayton S, Vadgama JV. Expression of FOXO1 is associated with GATA3 and Annexin-1 and predicts disease-free survival in breast cancer. Am J Cancer Res. 2012;2(1):104–115.
17. Bois PRJ, Izeradjene K, Houghton PJ, Cleveland JL, Houghton JA, Grosveld GC. FOXO1a acts as a selective tumor suppressor in alveolar rhabdomyosarcoma. J Cell Biol. 2005;170(6):903–912. doi:10.1083/jcb.200501040
18. Nahed BV, Redjal N, Brat DJ, et al. Management of patients with recurrence of diffuse low grade glioma: a systematic review and evidence-based clinical practice guideline. J Neurooncol. 2015;125(3):609–630. doi:10.1007/s11060-015-1910-2
19. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi:10.1016/j.tibs.2017.01.002
20. Li Z, Liu C, Huang C, et al. Quinazoline derivative QPB-15e stabilizes the c-myc promoter G-quadruplex and inhibits tumor growth in vivo. Oncotarget. 2016;7(23):34266–34276. doi:10.18632/oncotarget.9088
21. Masamha CP, Benbrook DM. Cyclin D1 degradation is sufficient to induce G1 cell cycle arrest despite constitutive expression of cyclin E2 in ovarian cancer cells. Cancer Res. 2009;69(16):6565–6572. doi:10.1158/0008-5472.CAN-09-0913
22. Li X, Bao C, Ma Z, et al. Perfluorooctanoic acid stimulates ovarian cancer cell migration, invasion via ERK/NF-κB/MMP-2/-9 pathway. Toxicol Lett. 2018;294:44–50. doi:10.1016/j.toxlet.2018.05.009
23. Rasool M, Malik A, Basit Ashraf MA, et al. Evaluation of matrix metalloproteinases, cytokines and their potential role in the development of ovarian cancer. PLoS One. 2016;11(11):e0167149. doi:10.1371/journal.pone.0167149
24. Wu ZS, Wu Q, Yang JH, et al. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int J Cancer. 2008;122(9):2050–2056. doi:10.1002/ijc.23337
25. Bertrand MJ, Milutinovic S, Dickson KM, et al. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol Cell. 2008;30(6):689–700. doi:10.1016/j.molcel.2008.05.014
26. Coomans de Brachène A, Demoulin J-B. FOXO transcription factors in cancer development and therapy. Cel Mol Life Sci. 2016;73(6):1159–1172. doi:10.1007/s00018-015-2112-y
27. Song H-M, Song J-L, Li D-F, Hua K-Y, Zhao B-K, Fang L. Inhibition of FOXO1 by small interfering RNA enhances proliferation and inhibits apoptosis of papillary thyroid carcinoma cells via Akt/FOXO1/Bim pathway. Onco Targets Ther. 2015;8:3565–3573. doi:10.2147/OTT.S95395
28. Kim SY, Yoon J, Ko YS, et al. Constitutive phosphorylation of the FOXO1 transcription factor in gastric cancer cells correlates with microvessel area and the expressions of angiogenesis-related molecules. BMC Cancer. 2011;11:264. doi:10.1186/1471-2407-11-264
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.