Back to Journals » OncoTargets and Therapy » Volume 12
TRIM3 Negatively Regulates Autophagy Through Promoting Degradation of Beclin1 in Ewing Sarcoma Cells
Authors Lu Q, Zhang Y, Ma L, Li D, Li M, Liu P, Li J
Received 17 June 2019
Accepted for publication 13 November 2019
Published 30 December 2019 Volume 2019:12 Pages 11587—11595
DOI https://doi.org/10.2147/OTT.S219777
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Arseniy Yuzhalin
Qunshan Lu, Yuankai Zhang, Liang Ma, Deqiang Li, Ming Li, Peilai Liu, Jianmin Li
Department of Orthopedics, Qilu Hospital of Shandong University, Jinan 250012, Shandong, People’s Republic of China
Correspondence: Peilai Liu; Jianmin Li
Department of Orthopedics, Qilu Hospital of Shandong University, Jinan 250012, Shandong, People’s Republic of China
Tel/fax +86 531-692-1941
Email [email protected]; [email protected]
Background and aim: Ewing sarcoma (ES) is an aggressive neoplasm predominantly occurring in adolescents and has a poor prognosis when metastasized. In the current study, we were aiming to investigate the function of TRIM3 in autophagy in ES cells.
Methods: The expression of TRIM3 in Ewing sarcoma tissues and normal tissues was examined by quantitative PCR and western blot. The effect of TRIM3 on autophagy was detected by western blot and immunofluorescence assay. Target of TRIM3 was examined by western blot, immunoprecipitation and ubiquitination assay.
Results: We found the expression of TRIM3 was significantly up-regulated in Ewing sarcoma tissues compared with normal tissues, and this phenomenon was regulated by EWS-FLI1 expression. Furthermore, we observed that overexpression of TRIM3 markedly and consistently inhibited autophagy in ES cells, and autophagy was enhanced in TRIM3-silenced ES cells. Finally, we found in ES cells, TRIM3 could directly interact with Beclin1, and improved its K48-linked polyubiquitinaion, leading to the degradation of Beclin1 and then regulated autophagy.
Conclusion: In the present research, for the first time we revealed that TRIM3 negatively regulates autophagy through promoting degradation of Beclin1 in Ewing sarcoma cells, and these findings may provide ideas for ES research.
Keywords: Ewing sarcoma, autophagy, TRIM3, Beclin1
Introduction
ES is an aggressive bone and soft tissue malignancy, which mainly affects children and young people.1 In recent years, with the development of multidrug systemic chemotherapy and active local control measures, the overall survival rate of patients with local diseases has been significantly improved.2 However, for the ~25% of patients who present with metastatic disease, the prognosis is poor and event-free survival rate for these patients remains <25%.3 Thus, it is of great significance to develop new novel therapeutic targets for the treatment of ES.
Autophagy is a eukaryotic homeostatic mechanism whereby cells remove from their cytoplasm toxic aggregates, damaged and surplus organelles, invading pathogens or utilize bulk cytosol for and metabolic needs.4 Abnormal autophagy has also been implicated in pathological development, emphasizing its critical involvement in maintaining homeostasis at the cellular and organismic level.5 Beclin1 (BECN1) is a B-cell lymphoma 2 (Bcl-2) homology 3 domain-only protein, it is a central protein that assembles cofactors for the formation of a BECN1-PIK3C3-PIK3R4 complex to trigger the autophagy protein cascade which are utilized in the initiation of autophagy.6 However, the effect of Beclin1 and its regulation in ES still remain largely unknown.
TRIM proteins share a similar characteristic structure, which includes a RING (R) domain, one or two B-boxes (B), and a coiled coil (CC) domain in the N-terminal and a domain in the C-terminal with variable structures.7,8 TRIM proteins are involved in a broad range of biological processes, including cell differentiation, apoptosis, transcriptional regulation, signal transduction, and immunity.9–11 Here, we identified a novel function for TRIM3 as an E3 ubiquitin ligase for Beclin1. For the first time, we found that TRIM3 expression is increased in Ewing sarcoma tissues and up-regulated by EWS-FLI1. TRIM3 was also found to suppress autophagy in ES cells and it directly interacted with Beclin1, promoting proteasomal degradation of Beclin1, therefore suppressed autophagy in ES cells. In conclusion, we revealed the effect of TRIM3 on autophagy in ES cells in the current study.
Materials and Methods
Cell Culture and Tissues
NIH3T3, A673, and TC71 cells were obtained from American Type Culture Collection (Manassas, VA). All the cells were cultured in RPMI 1640 containing 10% FCS for normal condition. Cells were cultured in serum-free medium for starvation for indicated time to induce autophagy. Eight formalin-fixed paraffin-embedded (FFPE) specimens of Ewing’s sarcomas and eight normal soft tissues around bones were acquired from Department of Orthopedics, Qilu Hospital of Shandong University, China. Clinical characteristics of ES patients were provided (Table 1). The study protocol was approved by the Ethics Committee of Our Hospital and all patients gave written informed consent.
 |
Table 1 Clinical Characteristics of ES Patients (n=8) |
Lentivirus and Plasmid
Lentivirus containing empty plasmid or EWS-FLI1 expression plasmid, TRIM3 expression plasmid, HA-tagged ub plasmids, shRNA-control plasmid and shRNA-TRIM3 plasmid were all constructed and bought from MDL biotechnology (MDL biotech, Beijing, China). Myc-tagged Beclin1 or Flag-tagged TRIM3 were obtained by PCR and cloned into the pCMV-Myc plasmid or pCMV6-Flag plasmid (Promega). The lentivirus particle was used to infect NIH3T3 for 3 days at 50 MOI with the presence of polybrene, puromycin selection was performed to establish the overexpression cell lines. Lipofectamine 2000 transfection reagent (Invitrogen) was used to transfect plasmids and siRNAs into ES cells.
RNA Analysis and ChIP Assays
The cells were collected in TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA was extracted using the TRIzol reagent according to the manufacturer’s instructions. A LightCycler (ABI PRISM 7000; Applied Biosciences) and a SYBR RT-PCR kit (Takara Biotechnology, Dalian, China) were used for real time PCR analysis. GAPDH was used as the internal control, thermocycling conditions were 1 cycle (95°C, 5 min) and 40 cycles (95°C, 15 sec; 57°C, 30 sec; 72°C, 30 sec) and the 2^-ΔΔCT method was used to evaluate the relative quantities of each amplified product in the samples.12 ChIP assay was performed as previously reported and the primer sequences for qPCR (as shown in Table 2) and ChIP-qPCR were used as described.13
 |
Table 2 List of Primers Used in the Study |
Western Blot Analysis and Immunoprecipitation
The cells were lysed using RIPA buffer (Thermo Scientific, Rockford, IL), and total protein in the supernatants was quantified using a BCA protein assay kit (Thermo Scientific, Waltham, MA). Immunoprecipitation (IP) and Western blot analysis were performed as described previously.14 Primary antibodies including anti-p62 (1:500, #sc-101542), anti-LC3 (1:500, #sc-398822), anti-FLI1 (1:500, #sc-22808) and anti-glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (1: 1000, #sc-32233) were bought from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA), anti-TRIM3 (1:1000, #ab111840), anti-Beclin1 (1:1000, #ab207612), anti-Myc (1: 1000, #ab32), anti-Flag (1:1000, #ab49763) and anti-HA (1:1000, #ab9110) were obtained from Abcam (Abcam, Cambridge, USA).
Confocal Microscopic Analysis
The experiment was performed as described previously.15 All slides were observed under a confocal laser microscopy (LSM780, Carl Zeiss, Oberkochen, Germany).
Statistical Analysis
All data are presented as mean ± S.D. of three independent experiments. Statistical significance was determined using the two-tailed student’s t-test to compare two groups. One-way ANOVA was performed to compare three or more groups. If the ANOVA analysis was significant, the Newman-Keuls test was applied for comparison between each two groups. Data were considered significant at P < 0.05.
Results
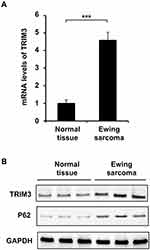
Expression of TRIM3 Was Increased in ES Tissues
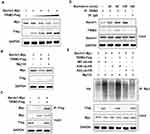
TRIM3 has been reported as an essential suppressor in glioblastomas, colorectal cancer and rheumatoid arthritis.16 In order to investigate the function of TRIM3 in ES, we examined the expression of TRIM3 in ES tissues at first. As shown in Figure 1A, the mRNA level of TRIM3 is significantly up-regulated in ES tissues compared with normal tissues. Consistently, the protein level of TRIM3 is also increased in ES tissues (Figure 1B). In addition, we also examined the expression of P62 which is well known as marker protein of autophagy, and we observed that P62 levels were also increased consistent with TRIM3 expression in ES tissues (Figure 1B). Taken together, these data suggested that expression of TRIM3 is increased in ES tissues.
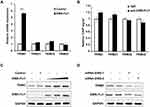
Expression of TRIM3 Was Regulated by EWS-FLI1
EWS-FLI1 induces the expression of many proteins, therefore we examined the relationship between TRIM3 and EWS-FLI1. As shown in Figure 2A, we detected the expression of various TRIM proteins including TRIM31, TRIM32, TRIM42, TRIM3, and only the expression of TRIM3 was markedly increased by EWS-FLI1 overexpression. EWS-FLI1 is known as a transcription factor to induce protein expression, we suspected if TRIM3 is a transcript target of EWS-FLI1. However, as shown in Figure 2B, ChIP-qPCR result demonstrated that EWS-FLI1 does not has the ability to interact with TRIM3 promoter, which meant that TRIM3 is not a transcript target of EWS-FLI1. Furthermore, we found the expression of TRIM3 was up-regulated in a dose dependent manner in EWS-FLI1 overexpressed cells (Figure 2C). Consistently, knockdown the expression of EWS-FLI1 also decreased TRIM3 expression. (Figure 2D). Taken together, these data suggested that TRIM3 expression is regulated by EWS-FLI1, however TRIM3 is not a transcript target of EWS-FLI1.
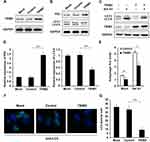
Overexpression of TRIM3 Inhibited Autophagy in ES Cells
In previous study, we found that EWS-FLI1 promotes autophagy in ES cells,13 therefore we supposed if TRIM3 could also regulate autophagy. Overexpression of TRIM3 in TC71 cells was confirmed as shown in Figure 3A. We observed that the expression of P62 was significantly increased and LC3-II level was decreased in TRIM3 overexpressed ES cells (Figure 3B and D). By using autophagy inhibitor Bafilomycin A1 (Baf A1), we observed that TRIM3 overexpression decreased autophagy flux (Figure 3C and E). Furthermore, we found that the formation of autophagosomes was suppressed by TRIM3 overexpression (Figure 3F and G). Taken together, these data suggested that overexpression of TRIM3 inhibits autophagy in ES cells.
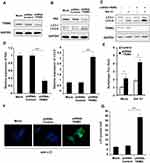
Silencing of TRIM3 Promoted Autophagy in ES Cells
ShRNA plasmid of TRIM3 was used to knockdown the expression of TRIM3, and the effect of shRNA was examined (Figure 4A). After transfection with shRNA-TRIM3, P62 level was greatly decreased and LC3-II expression was increased (Figure 4B and D), autophagy flux was enhanced (Figure 4C and E) as well as the increased formation of autophagosomes in TRIM3-silenced ES cells (Figure 4F and G). Taken together, these data suggested that silencing of TRIM3 promotes autophagy in ES cells.
TRIM3 Interacted with Beclin1 and Promoted K48-Linked Polyubiquitination of Beclin1
Beclin1 is a well-known key regulator of autophagy, we supposed if TRIM3 regulate autophagy through Beclin1. At first, we detected the expression of Beclin1 in TRIM3 overexpressed ES cells, and we found that TRIM3 significantly suppressed Beclin1 expression in a dose dependent manner and the decreased expression induced by TRIM3 was restored by Mg132 treatment (Figure 5A and B). We also examined the interaction between TRIM3 and Beclin1, and we observed that TRIM3 directly interacted with Beclin1 (Figure 5C). Endogenous interaction was also examined, and we found that TRIM3 expression is increased first and then decreased at 120mins after starvation treatment, the interaction between TRIM3 and Beclin1 is strongest at 120mins of starvation (Figure 5D). At last, we detected the level of polyubiquitination of Beclin1 in TRIM3 overexpressed ES cells, we found that overexpression TRIM3 markedly improved K48-linked polyubiquitination of Beclin1 but has no effect on level of K63-linked polyubiquitination (Figure 5E), therefore TRIM3 promoted the proteasomal degradation of Beclin1, leading to the suppression of autophagy in ES cells.
Discussion
ES is an aggressive bone and soft tissue malignant tumor that primarily affects children, and young adults.2 The survival rates for patients with metastatic disease have not improved in over 20 years.17 Thus, novel therapeutic targets and increased understanding of the metastatic mechanism of ES are required. Tripartite motif (TRIM) family proteins are composed of over 70 members in humans.18 TRIM proteins have been implicated in many biological processes including cell differentiation, apoptosis, transcriptional regulation, and signaling transduction.7,8 The characteristic structure of TRIM proteins is the presence of a RING (R) domain, one or two B-boxes (B), and a coiled coil (CC) domain.9 In the current study, we investigated the role of TRIM3 in ES. We found that the expression of TRIM3 is specific increased in ES tissues compared with the normal tissues. We also observed that the expression of TRIM3 is associated with EWS-FLI1 expression, however, TRIM3 was found not being a transcript target of EWS-FLI1, and the precise mechanism about EWS-FLI1 induced TRIM3 expression still need further study.
Autophagy plays great roles in the clearance of long-lived proteins, aggregates and damaged organelles, and dysregulation of autophagy is highly associated with cancer, particularly in tumorigenesis and chemotherapy resistance.13 In the current study, we found that TRIM3 could regulate autophagy in ES cells. Overexpression of TRIM3 significantly inhibits autophagy, as evidenced by the increases in the amount of P62 (SQSTM1) and decreases in the amount of LC3B-II, two important markers of autophagy, as well as the increased LC3 puncta in cells. Consistently, the level of P62 was up-regulated and LC3B-II was down-regulated in TRIM3-silenced ES cells. These findings indicated that TRIM3 plays great roles in the process of autophagy in ES.
To further explore the potential mechanisms that how could TRIM3 regulate autophagy in ES cells, we examined the relationship between TRIM3 and Beclin1, which has been demonstrated to initiate autophagosome formation.19 Beclin-1 dysfunction has been identified in a number of disorders, including cancer, aging and degenerative diseases; for example, overexpression of Beclin-1 markedly promotes autophagic cell death in leukemia cells.20 In the study, we found that the expression of Beclin1 is down-regulated by TRIM3 overexpression in a dose-dependent manner. The interaction between TRIM3 and Beclin1 was also proved by IP assay, indicated that TRIM3 directly interact with Beclin1. Most importantly, we found that the K48-linked polyubiquitination level of Beclin1 was increased in TRIM3 overexpressed ES cells, which meant that TRIM3 increased K48-linked polyubiquitination of Beclin1 and promoted Beclin1 degradation, and finally down-regulated the autophagy in ES cells.
In conclusion, in the present research, for the first time we revealed the function of TRIM3 in ES and we showed the effect of TRIM3 on autophagy in ES cells, and these findings may provide ideals for ES researches.
Abbreviations
ES, Ewing sarcoma; TRIM3, Tripartite motif-containing 3; ChIP, Chromatin Immunoprecipitation.
Acknowledgements
This study was supported by grants from the Natural Science Foundation of Shandong Province (No. ZR2015HQ018) and China Postdoctoral Science Foundation Grant (No. 2018M632680).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.v61:2
2. Balamuth NJ, Womer RB. Ewing’s sarcoma. Lancet Oncol. 2010;11(2):184–192. doi:10.1016/S1470-2045(09)70286-4
3. Gorlick R, Janeway K, Lessnick S, Randall RL, Marina N, Committee COGBT. Children’s oncology group’s 2013 blueprint for research: bone tumors. Pediatr Blood Cancer. 2013;60(6):1009–1015. doi:10.1002/pbc.v60.6
4. Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi:10.1146/annurev-cellbio-092910-154005
5. Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. N Engl J Med. 2013;368(19):1845–1846. doi:10.1056/NEJMra1205406
6. Oberstein A, Jeffrey PD, Shi Y. Crystal structure of the Bcl-XL-beclin 1 peptide complex: beclin 1 is a novel BH3-only protein. J Biol Chem. 2007;282(17):13123–13132. doi:10.1074/jbc.M700492200
7. Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27(11):1147–1157. doi:10.1002/(ISSN)1521-1878
8. Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3(10):799–808. doi:10.1038/nrmicro1248
9. Ozato K, Shin DM, Chang TH, Morse HC
10. Kawai T, Akira S. Regulation of innate immune signalling pathways by the tripartite motif (TRIM) family proteins. EMBO Mol Med. 2011;3(9):513–527. doi:10.1002/emmm.v3.9
11. McNab FW, Rajsbaum R, Stoye JP, O’Garra A. Tripartite-motif proteins and innate immune regulation. Curr Opin Immunol. 2011;23(1):46–56. doi:10.1016/j.coi.2010.10.021
12. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408.
13. Lu Q, Zhang Y, Ma L, et al. EWS-FLI1 positively regulates autophagy by increasing ATG4B expression in ewing sarcoma cells. Int J Mol Med. 2017;40(4):1217–1225. doi:10.3892/ijmm.2017.3112
14. Lin Y, Luo Z. NLRP6 facilitates the interaction between TAB2/3 and TRIM38 in rheumatoid arthritis fibroblast-like synoviocytes. FEBS Lett. 2017;591(8):1141–1149. doi:10.1002/1873-3468.12622
15. Guo L, Huang JX, Liu Y, et al. Transactivation of Atg4b by C/EBPbeta promotes autophagy to facilitate adipogenesis. Mol Cell Biol. 2013;33(16):3180–3190. doi:10.1128/MCB.00193-13
16. Wang M, Wu J, Guo Y, Chang X, Cheng T. The tripartite motif-containing protein 3 on the proliferation and cytokine secretion of rheumatoid arthritis fibroblast-like synoviocytes. Mol Med Rep. 2017;15(4):1607–1612. doi:10.3892/mmr.2017.6164
17. Zhou Z, Yu L, Kleinerman ES. EWS-FLI-1 regulates the neuronal repressor gene REST, which controls ewing sarcoma growth and vascular morphology. Cancer. 2014;120(4):579–588. doi:10.1002/cncr.28555
18. Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. doi:10.1186/1471-2148-8-225
19. Kang R, Zeh HJ, Lotze MT, Tang D. The beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011;18(4):571–580. doi:10.1038/cdd.2010.191
20. Tong Y, You L, Liu H, et al. Potent antitumor activity of oncolytic adenovirus expressing beclin-1 via induction of autophagic cell death in leukemia. Oncotarget. 2013;4(6):860–874. doi:10.18632/oncotarget.v4i6
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.