Back to Journals » Cancer Management and Research » Volume 12
TRIB3 Promotes Lung Adenocarcinoma Progression via an Enhanced Warburg Effect
Authors Xing Y, Luo P, Hu R, Wang D, Zhou G, Jiang J
Received 23 October 2020
Accepted for publication 10 December 2020
Published 23 December 2020 Volume 2020:12 Pages 13195—13206
DOI https://doi.org/10.2147/CMAR.S287956
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yutong Xing,1,2,* Peng Luo,2,* Rui Hu,1 Duanduan Wang,1 Gang Zhou,2 Jie Jiang3
1Department of Cardiothoracic Surgery, The Fifth Hospital of Xiamen, Xiamen, People’s Republic of China; 2Department of Cardiothoracic Surgery, The First Affiliated Hospital of Jiamusi University, Jiamusi, People’s Republic of China; 3Department of Thoracic Surgery, The First Affiliated Hospital of Xiamen University, Xiamen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yutong Xing Department of Cardiothoracic Surgery
The Fifth Hospital of Xiamen, Xiamen, Fujian 361101, People’s Republic of China
Email [email protected]
Background: The pseudokinase Tribbles 3 (TRIB3) is involved in many cellular processes and various cancers. In recent years, the importance of metabolic transformation in the maintenance of malignant tumors has become increasingly prominent. Abnormal metabolism of cancer cells is considered a hallmark of cancer. However, the exact role and molecular mechanism of TRIB3 in lung adenocarcinoma (LUAD) cell reprogramming is largely unknown.
Methods: The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) of cells were examined with a Seahorse XF Extracellular Flux Analyzer. In vitro and in vivo RT-qPCR, Western blotting, and functional assays were performed to explore the functional roles of TRIB3 in LUAD.
Results: In the present study, we demonstrated that TRIB3 is remarkably upregulated in LUAD cell lines as well as tissues. TRIB3 knockdown significantly inhibited LUAD cell growth and suppressed LUAD cell invasion, while TRIB3 overexpression conferred the opposite effects. Moreover, silencing TRIB3 suppressed the tumorigenesis and metastatic ability of LUAD cells. Mechanistically, we demonstrated that silencing TRIB3 significantly impaired aerobic glycolysis ability in LUAD cells. Furthermore, our data indicated that TRIB3 knockdown decreased hypoxia-inducible factor (HIF)1α levels and targeted the glycolytic genes regulated by HIF1α.
Conclusion: Together, our findings revealed a previously unappreciated function of TRIB3 in cancer cell metabolism and tumor progression, illustrating that TRIB3 could be considered a valuable therapeutic target for LUAD patients.
Keywords: TRIB3, LUAD, aerobic glycolysis, HIF1α
Background
Lung cancer remains an aggressive malignant disease with poor prognosis.1 Non-small-cell lung cancer (NSCLC) accounts for nearly 80% of lung cancers, most of which are LUAD. Early diagnosis of LUAD has been significantly improved by newly developed treatments, but the 5-year survival rate of patients with LUAD is still relatively low.2 Therefore, it is of great importance to explore the molecular mechanism of the occurrence and development of LUAD and to explore new targets for the treatment of LUAD.
An emerging hallmark of cancer cells is metabolic alterations, with most, if not all, cancer cells producing energy primarily through glycolysis in the cytosol.3–6 Otto Warburg published a series of studies demonstrating that cancer cells exhibit metabolic characteristics that are different from those of normal tissues, which is also known as aerobic glycolysis or the Warburg effect.7 It is now clear that in many types of cancer cells, tumor formation is largely due to enhanced aerobic glycolysis. For example, hypoxia-inducible factor 1 (HIF-1) has been suggested to be responsible for the increased transcription of genes encoding glycolytic enzymes and glucose transporters, including PDK1 (pyruvate dehydrogenase kinase, isoenzyme 1) and LDHA (lactate dehydrogenase A).8–10 The oncogene cMyc is also involved in the regulation of many glycolysis-related genes, including LDHA and GLUT1 (glucose transporter 1).11–13 These studies indicate that metabolic reprogramming of cancer cells is much more complicated than expected and deserves further study.
The pseudokinase Tribble 3 (TRIB3), as a pressure sensor, responds to a variety of different stressors and participates in chronic inflammation, metabolism and malignant diseases.14 An increasing number of studies have shown that TRIB3 can be increased by multiple stimuli and regulate the signaling pathways of transforming growth factor-β, phosphatidylinositol 3-kinase (PI3K), and mitogen-activated protein kinase (MAPK), thereby participating in the glucose/lipid metabolism, cell differentiation and cell survival of tumor cells.15 Recently, several groups have found TRIB3 to be a key factor regulating tumorigenesis and tumor progression. In particular, TRIB3 is upregulated in various cancer tissues and closely related to the poor prognosis of patients, including breast cancer. Recent studies have demonstrated that TRIB3 is a key factor in regulating tumorigenesis and development. In particular, TRIB3 is upregulated in various tumor tissues and is closely related to the poor prognosis of colorectal cancer16 and breast cancer.17 However, as one of the new hallmarks of LUAD, the role of TRIB3 in the metabolic reprogramming of cancer cells has rarely been studied.
In this study, our data confirmed the effect of TRIB3 on the malignancy of LUAD. TRIB3 promoted LUAD cell proliferation and invasion by inducing aerobic glycolysis. TRIB3 caused aerobic glycolysis and inhibited HIF1α protein levels. In conclusion, our current study reveals a new role of TRIB3 in regulating tumor progression and aerobic glycolysis in LUAD.
Method
Cell Lines and Cell Culture
The LUAD cell line and epithelial cell line 16HBE were purchased from the Cell Bank of Type Culture Collection of the Shanghai Institute of Cell Biology, Chinese Academy of Sciences. H1975, A549, H226 and H1299 cells were cultured in RPMI-1640 (Thermo, USA) containing 10% fetal bovine serum (FBS), and 16HBE and SPC-A1 cells were cultured in DMEM (Thermo, USA) containing 10% FBS. In addition, streptomycin (100 U/mL) and penicillin (100 U/mL) were added to all cell culture media at 37 °C in a 5% CO2 incubator.
Patients and Tissue Specimens
Forty-two pairs of LUAD tumor samples as well as corresponding adjacent normal tissues were collected from June 2018 to January 2020 at the First Affiliated Hospital of Jiamusi University. Samples were rapidly frozen and stored at −80 °C until use in qRT-PCR experiments. The study was approved by the Research Ethics Committee of Jiamusi University and was consistent with the Declaration of Helsinki. All patients involved in the experiment provided written informed consent.
Reverse Transcription and Real-Time Quantitative PCR (RT-qPCR)
We used TRIzol to isolate total RNA under the guidance provided and agarose gel electrophoresis to assess the integrity of RNA. cDNA was synthesized from 1 μg total RNA through the superscript III first-strand synthesis system (Toyobo, Osaka, Shiga, Japan) using 10 μL volumes with the following thermocycler settings: 37 °C for 15 minutes, 50 °C for 5 minutes, and 98 °C for 5 minutes. Fast SYBR Green Master Mix (Applied Biosystems Inc. CA, USA) for qRT-PCR. Primer sequences are listed in Supplementary Table 1.
Plasmid Construction, Lentivirus Production and Stable Cell Line Selection
We used lentivirus-mediated transfection to silence TRIB3 expression. The pLKO.1 TRC cloning vector was used to construct shRNA against TRIB3. The targets for TRIB3 were 5ʹ-CAGGAAGAAGCGGTTGGAGTTGGAT-3ʹ and 5ʹ-AGGGAGACAGAGAAGTGGTTCTGTA-3ʹ. A549 cells were infected with lentiviral particles, and then puromycin selection was applied to obtain the stable cell line. TRIB3 was cloned into the pcDNA3.1(+) (Invitrogen) vector to obtain TRIB3 overexpression constructs. Cells (2x105/well) were seeded into 6-well plates. At 60–70% fusion, cells were transfected with 4 μg of plasmid using Lipofectamine 2000 according to the manufacturer’s protocol. Thirty-six hours after transfection, cells were collected for subsequent experiments.
CCK-8 Assay
Cell viability was examined with the Cell Counting Kit 8 (CCK-8) according to the manufacturer’s protocol (Dojindo, Japan). Cells (2μ103 cells/) were transfected with siRNAs or plasmids in 96-well plates 24 h after seeding and then cultured for 2 days. The medium in each well was replaced with 100 μL complete medium containing 10 μL CCK-8 solution. Absorbance at 450 nm was measured with Doscan spectroscopy (Thermo Fisher, Rockford, IL, USA).
Colony Formation Assay
Cells were seeded in 6-well plates (103/well) with different treatments. The colonies were treated with methanol followed by staining with 0.1% crystal violet after two weeks (Sigma-Aldrich). The number and statistical analysis of visible colonies were described previously.18
In vivo Tumor Growth Model
LUAD cells were stably transfected with sh-TRIB3 or scrambled control vector, harvested, washed with PBS, and resuspended at a concentration of 1 μ 108 cells/mL. Each mouse was subcutaneously injected with 100 μL suspension cells unilaterally into the hind abdomen. We examined tumor growth every 5 days and calculated tumor volume. After 30 days, mice were sacrificed according to the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals, and tumors were isolated for measurement and weighing. The protocol was approved by the Ethics Committee of Animal Experiments of Xiamen University.
In vivo Lung-Colonization Assays
A549 cells (2 x 106 cells per mouse) were injected into the tail vein of nude mice. Lungs were collected at week 5 and fixed with 10% formalin as previously described.19 The number of metastatic colonies on the lung surface was calculated under an anatomical microscope. All animal experiments were approved by the Animal Care Committee of Xiamen University. The lungs were dissected, placed in liquid nitrogen or 4% formalin, and fixed at temperature for 30 minutes. Next, 5-µm-thick sections were scanned with a Scanscope XT digital slide scanner (Aperio Technologies, Inc.) with hematoxylin and eosin staining for 10 minutes at room temperature. Digital images of lung sections were analyzed for metastatic burden. Lung tumor lesions were digitally calibrated using Spectrum software (version 11.2.0.3; Aperio Technologies, Inc.). The burden of metastasis was calculated as the average of four-step sections of the total area of the tumor lesion divided by the total area of the lung.
Measurement of the Oxygen Consumption Rate (OCR) and Extracellular Acidification Rate (ECAR)
We examined the cellular glycolytic capacity as well as mitochondrial function with a Seahorse Bioscience Extracellular Flux Analyzer according to the manufacturer’s instructions (Seahorse Bioscience, Billerica, MA, USA). One day before the assay, cells were plated in cell culture microplates (Hippocampal Biosciences). The Seahorse buffer consisted of DMEM, phenol red, 25 mM glucose, 2 mM sodium pyruvate and 2 mM glutamine. When ECAR values were measured, 10 mM glucose, 1 μM oligomycin and 100 mM 2-deoxyglucose were added automatically. After monitoring baseline respiration, 1 μM oligomycin, 1 M FCCP and 1 μM rotenone were automatically injected into cell culture microplates to measure OCR. ECAR and OCR values were calculated as described previously.20
Statistical Analysis
GraphPad Prism 7.0 was used to perform Student’s t-test, ANOVAs, or repeated-measures ANOVAs to assess the statistical significance of differences between two or more groups. All data were expressed as the mean ± the SEM. P < 0.05 was considered statistically significant.
Results
TRIB3 Levels are Upregulated in LUAD Tissue and Cell Lines
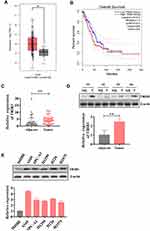
First, we evaluated TRIB3 expression from The Cancer Genome Atlas (TCGA) database of LUAD samples and adjacent normal samples. As shown in Figure 1A, TRIB3 was remarkably upregulated in LUAD tissues compared with adjacent normal tissues. In terms of overall survival, patients with high expression of TRIB3 had significantly poorer prognoses than those with low expression of TRIB3 (Figure 1B). These results suggest that TRIB3 overexpression may contribute to the development of new prognostic or progression markers in lung adenocarcinoma.
Then, we examined the expression of TRIB3 in primary LUAD patients and adjacent normal lung tissues by RT-qPCR as well as Western blot, and the results showed that TRIB3 expression in tumor tissues was significantly higher than that in paired control tissues (Figure 1C and D). We further detected the expression of TRIB3 in five lung cancer cell lines (A549, SPC-A1, H1299, H226 and H1975) and the human bronchial epithelial cell line 16HBE by RT-qPCR. LUAD cells showed a significant level of TRIB3 expression compared with 16HBE cells (Figure 1E). Overall, these results indicate that TRIB3 is upregulated in LUAD, suggesting that it may be associated with the progression of LUAD.
Knockdown of TRIB3 Suppresses LUAD Cell Growth and Invasion
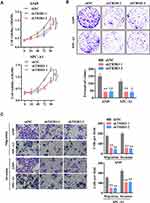
To directly assess the possible role of TRIB3 in LUAD, LUAD cell lines A549 and SPC-A1 were transfected with two specific shRNAs targeting TRIB3, both of which effectively silenced the expression of endogenous TMPO-AS1 (data not shown). The CCK-8 assay showed that TRIB3 knockdown significantly inhibited the growth of LUAD cells (Figure 2A). Consistent with the results of the CCK-8 assay, the colony formation assay confirmed that TRIB3 knockdown significantly suppressed the proliferation of LUAD cells, forming fewer and smaller colonies in soft agar (Figure 2B). These results suggest that TRIB3 has a growth-promoting effect in vitro. To further determine whether TRIB3 is associated with the progression of LUAD, we analyzed the effects of TRIB3 inhibition on the invasive behavior of A549 and SPC-A1 cells. Our Transwell invasion assay data showed that shRNA-mediated downregulation of TRIB3 significantly inhibited the invasive ability of LUAD cells in A549 and SPC-A1 cells (Figure 2C). In conclusion, these data indicate that TRIB3 plays an essential role in the development of LUAD.
TRIB3 Overexpression Promotes the Proliferation and Migration of LUAD Cells
We further explored the functional effect of pcDNA3.1-TRIB3 plasmid-mediated TRIB3 overexpression in LUAD cells. After 36 h of transfection, the expression of TRIB3 was significantly increased in A549 and SPC-A1 cells transfected with the pcDNA3.1-TRIB3 plasmid compared with those transfected the pcDNA3.1 vector alone (data not shown). CCK-8 experiments showed that TRIB3 upregulation promoted the proliferation of both types of cells (Figure 3A). In addition, in the TRIB3 overexpression group, the number of colonies was significantly higher (Figure 3B). Furthermore, increased TRIB3 expression promoted the invasion of A549 and SPC-A1 cells (Figure 3C and D). Taken together, these results indicate that the overexpression of TRIB3 promotes the progression of LUAD.
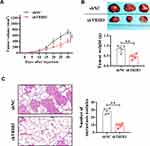
TRIB3 Promotes Tumor Growth and Metastasis in LUAD in vivo
With a xenograft mouse model, we explored the impact of TRIB3 inhibition on LUAD growth in vivo. A549 cells stably transfected with sh-TRIB3 or empty vector were injected into nude mice. After four weeks, we found that mice injected with A549 cells transfected with sh-TRIB3 showed significantly reduced tumor volume and tumor weight compared with mice of the control group (Figure 4A and B). Next, we evaluated the physiological relevance of TRIB3 to LUAD metastasis in vivo. TRIB3-downregulated A549 cells were injected into the tail vein of BALB/c nude mice. HE staining of the lungs after 4 weeks showed that sh-TRIB3 inhibited the occupation of LUAD in the lungs (Figure 4C). Taken together, we discovered that sh-TRIB3 effectively inhibited tumor growth and metastasis in LUAD in vivo.
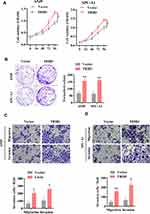
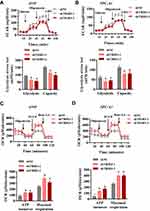
TRIB3 Promotes Aerobic Glycolysis in LUAD Cells
It is generally believed that proliferating solid cancer cells shift their glycometabolic pattern to hypoxic glycolysis. Because of the positive role of TRIB3 in predicting the prognosis and proliferation of tumor cells, we speculated that TRIB3 would be involved in regulating aerobic glycolysis. We first examined the effect of TRIB3 knockdown on glycolysis using a Seahorse XF extracellular flux analyzer. Our results suggested that ECAR decreased significantly in TRIB3-silenced A549 and SPC-A1 cells, reflecting the positive role of TRIB3 glycolysis in LUAD cells (Figure 5A and B). Cellular oxygen consumption reflects mitochondrial respiration. During aerobic glycolysis, cellular OCR decreases. Consistent with the ECAR results, we observed that TRIB3 knockdown increased the OCR in A549 and SPC-A1 cells, which reinforced the positive effect of aerobic glycolysis (Figure 5C and D). Therefore, TRIB3 positively regulates aerobic glycolysis in LUAD cells.
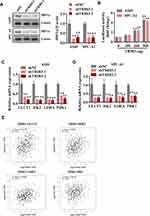
TRIB3 Positively Regulates HIF1α Protein Levels and Transcriptional Activity
As a main regulator of aerobic glycolysis, HIF1α plays an essential role in hypoxia adaptation in solid tumors. To assess whether TRIB3 regulates aerobic glycolysis by regulating HIF1α, we first measured HIF1α protein levels in TRIB3-silenced LUAD cells. As shown in Figure 6A, HIF1α protein levels decreased significantly when TRIB3 expression was inhibited. Next, we evaluated the effect of TRIB3 on the transcriptional activity of HIF1α as reflected by HRE-luciferase activity. Our data demonstrated that TRIB3 positively regulated the transcriptional activity of HIF1α in a dose-dependent manner, which reflected the positive role of TRIB3 in the regulation of the receptor pathway of HIF1α (Figure 6B). HIF1α regulates aerobic glycolysis by regulating the transcription of many glycolytic genes, including GLUT1, HK2, LDHA and PDK1. Therefore, the expression levels of these glycolytic genes were examined in TRIB3-silenced LUAD cells. Consistent with the glycolysis analysis results, GLUT1, HK2, LDHA and PDK1 decreased in TRIB3-silenced LUAD cells (Figure 6C and D). In addition, data from the TCGA database were analyzed by the GEPIA (Gene Expression Profiling Interactive Analysis) platform21 to determine the correlation between TRIB3 and glycolytic enzymes at the mRNA level in clinical LUAD specimens. Our analysis showed that TRIB3 was positively correlated with GLUT1, HK2, LDHA and PDK1 (Figure 6E).
Discussion
In this study, through in vitro and in vivo analysis, we found that TRIB3 is a tumor promoter in LUAD. TRIB3 was upregulated in LUAD patient samples and LUAD cell lines, suggesting that TRIB3 may play a role as a tumor regulator and may have value as a therapeutic target. In addition, the overexpression of TRIB3 promoted cell growth and invasion, while the downregulation of TRIB3 in LUAD cells resulted in suppressed cell proliferation and invasion. TRIB3 also demonstrated its growth-promoting and prometastatic effects in xenograft mouse models. In recent years, the progression and importance of metabolic reprogramming in tumorigenesis and development have become increasingly prominent, and metabolic reprogramming is considered one of the important markers of tumorigenesis and development.22,23 Mechanistic studies have shown that TRIB3 positively regulates aerobic glycolysis by regulating hypoxia-inducible factor (HIF) 1α, which has rarely been discussed before. Our results suggest that TRIB3 is a potential new therapeutic target for the treatment of LUAD.
As a stress sensor, pseudokinase Tribble 3 (TRIB3) responds to multiple stressors and participates in chronic inflammation, metabolism and malignant diseases by interacting with signal transduction proteins and functional proteins.14,24 A recent study reported that TRIB3 impairs the degradation function of autophagy and the proteasome by interacting with the autophagy receptor p62, thus promoting the occurrence and development of some cancers. Autophagy and the proteasome are two important protein quality control systems in cancer cells. The deletion of TRIB3 results in a significant reduction in the expression of tumor-promoting factors, including EGFR.25,26 TCGA data showed 1.14% gene amplification and 1.84% gene mutation for TRIB3 in NSCLC. Also, growth arrest-specific 5 (GAS5) could bind to TRIB3 protein to promote the degradation of TRIB3 protein and inhibited the high glucose-induced proliferation, anti-apoptosis, and migration of NSCLC cells.27 Moreover, TRIB3 has a potentially carcinogenic role in LUAD by binding to ERK and JNK and promoting the phosphorylation of ERK and JNK.28 In our study, the TRIB3 gene was expressed at higher levels in LUAD cell lines and the cancer tissues of LUAD patients than in healthy controls. Based on these results, we chose the TRIB3 gene as a treatment target in these experiments. This study revealed that TRIB3 plays a key role in promoting the occurrence and development of LUAD.
The microenvironment in which solid tumor cells reside is remote from blood vessels, resulting in oxygen supply and restricted nutrients; thus, tumor cells have evolved an ability to survive well in a hypoxic environment, which we call hypoxic adaptation.29 The most characteristic metabolic reprogramming in cancer is aerobic glycolysis, and the result of this hypoxic adaptation provides a metabolic advantage for cancer cells to proliferate and metastasize. However, few studies have reported the relationship between TRIB3 and aerobic glycolysis. Our study shows for the first time that TRIB3 acts as a positive regulator in the process of aerobic glycolysis in LUAD. Through the analysis of TCGA, we also confirmed the positive regulation of glycolytic genes by TRIB3, which was further confirmed in the in vitro experiments of LUAD cell lines. HIF1α plays a central role in hypoxic adaptation and has received extensive attention since its discovery.30 A series of posttranscriptional modifications of HIF1α, including hydroxylation, phosphorylation, acetylation and ubiquitination, are involved in the regulation of its activity.31,32 Our data demonstrate that TRIB3 regulates the expression and activity of HIF1α and further regulates glycolysis. However, there are still some issues to be explored, such as the changes in the expression of TRIB3 under hypoxic conditions and whether hypoxia epigenetically regulates TRIB3 and its underlying molecular basis.
Conclusion
Overall, our study uncovered a novel role of TRIB3 in the regulation of tumor progression and aerobic glycolysis in LUAD.
Ethics Approval
Ethical approval for this study was obtained from the Ethics Committee of Jiamusi University.
Funding
This work was supported by the Heilongjiang Health Department Fund [grant number: 2020-346].
Disclosure
The authors declare that they have no competing interests or conflicts of interest for this work.
References
1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi:10.3322/caac.21166
2. Martin P, Leighl NB. Review of the use of pretest probability for molecular testing in non-small cell lung cancer and overview of new mutations that may affect clinical practice. Ther Adv Med Oncol. 2017;9(6):405–414.
3. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674.
4. Compan V, Pierredon S, Vanderperre B, et al. Monitoring mitochondrial pyruvate carrier activity in real time using a bret-based biosensor: investigation of the warburg effect. Mol Cell. 2015;59(3):491–501.
5. Lin L, Huang H, Liao W, et al. MACC1 supports human gastric cancer growth under metabolic stress by enhancing the Warburg effect. Oncogene. 2015;34(21):2700–2710.
6. Iansante V, Choy PM, Fung SW, et al. PARP14 promotes the Warburg effect in hepatocellular carcinoma by inhibiting JNK1-dependent PKM2 phosphorylation and activation. Nat Commun. 2015;6:7882.
7. Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033.
8. Makanji Y, Tagler D, Pahnke J, Shea LD, Woodruff TK. Hypoxia-mediated carbohydrate metabolism and transport promote early-stage murine follicle growth and survival. Am J Physiol Endocrinol Metab. 2014;306(8):E893–903.
9. Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269(38):23757–23763.
10. Kim J-W, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3(3):177–185. doi:10.1016/j.cmet.2006.02.002
11. Osthus RC, Shim H, Kim S, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275(29):21797–21800. doi:10.1074/jbc.C000023200
12. Shim H, Dolde C, Lewis BC, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci U S A. 1997;94(13):6658–6663.
13. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47.
14. Mondal D, Mathur A, Chandra PK. Tripping on TRIB3 at the junction of health, metabolic dysfunction and cancer. Biochimie. 2016;124:34–52.
15. Yokoyama T, Nakamura T. Tribbles in disease: signaling pathways important for cellular function and neoplastic transformation. Cancer Sci. 2011;102(6):1115–1122.
16. Miyoshi N, Ishii H, Mimori K, et al. Abnormal expression of TRIB3 in colorectal cancer: a novel marker for prognosis. Br J Cancer. 2009;101(10):1664–1670.
17. Wennemers M, Bussink J, Scheijen B, et al. Tribbles homolog 3 denotes a poor prognosis in breast cancer and is involved in hypoxia response. Breast Cancer Research. 2011;13(4):R82.
18. Mu X, Wu H, Liu J, et al. Long noncoding RNA TMPO-AS1 promotes lung adenocarcinoma progression and is negatively regulated by miR-383-5p. Biomed Pharmacother. 2020;125:109989.
19. Chen Y, Deng X, Chen W, et al. Silencing of microRNA-708 promotes cell growth and epithelial-to-mesenchymal transition by activating the SPHK2/AKT/beta-catenin pathway in glioma. Cell Death Dis. 2019;10(6):448.
20. Tang J, Yan T, Bao Y, et al. LncRNA GLCC1 promotes colorectal carcinogenesis and glucose metabolism by stabilizing c-Myc. Nat Commun. 2019;10(1):3499.
21. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102.
22. Elia I, Doglioni G, Fendt SM. Metabolic Hallmarks of Metastasis Formation. Trends Cell Biol. 2018;28(8):673–684.
23. Zielinski DC, Jamshidi N, Corbett AJ, Bordbar A, Thomas A, Palsson BO. Systems biology analysis of drivers underlying hallmarks of cancer cell metabolism. Sci Rep. 2017;7:41241.
24. Schwarzer R, Dames S, Tondera D, Klippel A, Kaufmann J. TRB3 is a PI 3-kinase dependent indicator for nutrient starvation. Cell Signal. 2006;18(6):899–909.
25. Hua F, Li K, Yu JJ, et al. TRB3 links insulin/IGF to tumour promotion by interacting with p62 and impeding autophagic/proteasomal degradations. Nat Commun. 2015;6:7951.
26. Wang XJ, Li FF, Zhang YJ, Jiang M, Ren WH. TRIB3 promotes hepatocellular carcinoma growth and predicts poor prognosis. Cancer Biomark. 2020.
27. Ding CZ, Guo XF, Wang GL, et al. High glucose contributes to the proliferation and migration of non-small cell lung cancer cells via GAS5-TRIB3 axis. Biosci Rep. 2018.
28. Cao X, Fang X, Malik WS, et al. TRB3 interacts with ERK and JNK and contributes to the proliferation, apoptosis, and migration of lung adenocarcinoma cells. J Cell Physiol. 2020;235(1):538–547.
29. Rankin EB, Giaccia AJ. The role of hypoxia-inducible factors in tumorigenesis. Cell Death Differ. 2008;15(4):678–685.
30. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337.
31. Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128.
32. Bertout JA, Patel SA, Simon MC. The impact of O2 availability on human cancer. Nat Rev Cancer. 2008;8(12):967–975.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.