Back to Journals » Infection and Drug Resistance » Volume 13
Trends in Molecular Markers Associated with Resistance to Sulfadoxine-Pyrimethamine (SP) Among Plasmodium falciparum Isolates on Bioko Island, Equatorial Guinea: 2011–2017
Authors Lin LY, Li J, Huang HY, Liang XY, Jiang TT, Chen JT, Ehapo CS, Eyi UM, Zheng YZ, Zha GC, Xie DD, Wang YL, Chen WZ, Liu XZ, Lin M
Received 1 November 2019
Accepted for publication 19 March 2020
Published 28 April 2020 Volume 2020:13 Pages 1203—1212
DOI https://doi.org/10.2147/IDR.S236898
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Li-Yun Lin,1– 3,* Jian Li,4,* Hui-Ying Huang,5,6 Xue-Yan Liang,5,6 Ting-Ting Jiang,4 Jiang-Tao Chen,7,8 Carlos Salas Ehapo,9 Urbano Monsuy Eyi,9 Yu-Zhong Zheng,1 Guang-Cai Zha,1 Dong-De Xie,7,8 Yu-Ling Wang,7,8 Wei-Zhong Chen,5,6 Xiang-Zhi Liu,5,6 Min Lin1,5,6
1School of Food Engineering and Biotechnology, Hanshan Normal University, Chaozhou, Guangdong Province, People’s Republic of China; 2University of Chinese Academy of Sciences, Beijing, People’s Republic of China; 3CAS Key Laboratory of Tropical Marine Bio-Resources and Ecology, Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou, People’s Republic of China; 4Department of Human Parasitology, School of Basic Medical Sciences; Department of Infectious Diseases, Renmin Hospital, Hubei University of Medicine, Shiyan, Hubei Province, People’s Republic of China; 5Department of Medical Laboratory, Chaozhou People’s Hospital Affiliated to Shantou University Medical College, Chaozhou, Guangdong Province, People’s Republic of China; 6Department of Medical Genetics, Shantou University Medical College, Shantou, Guangdong Province, People’s Republic of China; 7The Chinese Medical Aid Team to the Republic of Equatorial Guinea, Guangzhou, Guangdong Province, People’s Republic of China; 8Department of Medical Laboratory, Huizhou Central Hospital, Huizhou, Guangdong Province, People’s Republic of China; 9Department of Medical Laboratory, Malabo Regional Hospital, Malabo, Equatorial Guinea
*These authors contributed equally to this work
Correspondence: Min Lin
School of Food Engineering and Biotechnology, Hanshan Normal University, Chaozhou, Guangdong Province, People’s Republic of China
Tel/Fax +86 768-2317422
Email [email protected]
Purpose: Antimalarial drug resistance is one of the major challenges in global efforts to control and eliminate malaria. In 2006, sulfadoxine-pyrimethamine (SP) replaced with artemisinin-based combination therapy (ACT) on Bioko Island, Equatorial Guinea, in response to increasing SP resistance, which is associated with mutations in the dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps) genes.
Patients and Methods: To evaluate the trend of molecular markers associated with SP resistance on Bioko Island from 2011 to 2017, 179 samples collected during active case detection were analysed by PCR and DNA sequencing.
Results: Pfdhfr and Pfdhps gene sequences were obtained for 90.5% (162/179) and 77.1% (138/179) of the samples, respectively. For Pfdhfr, 97.5% (158/162), 95.7% (155/162) and 98.1% (159/162) of the samples contained N51I, C59R and S108N mutant alleles, respectively. And Pfdhps S436A, A437G, K540E, A581G, and A613S mutations were observed in 25.4% (35/138), 88.4% (122/138), 5.1% (7/138), 1.4% (2/138), and 7.2% (10/138) of the samples, respectively. Two classes of previously described Pfdhfr-Pfdhps haplotypes associated with SP resistance and their frequencies were identified: partial (IRNI-SGKAA, 59.4%) and full (IRNI-SGEAA, 5.5%) resistance. Although no significant difference was observed in different time periods (p> 0.05), our study confirmed a slowly increasing trend of the frequencies of these SP-resistance markers in Bioko parasites over the 7 years investigated.
Conclusion: The findings reveal the general existence of SP-resistance markers on Bioko Island even after the replacement of SP as a first-line treatment for uncomplicated malaria. Continuous molecular monitoring and additional control efforts in the region are urgently needed.
Keywords: malaria, drug resistance, dihydrofolate reductase, dihydropteroate synthase, sulfadoxine-pyrimethamine
Introduction
According to the World Malaria Report 2019,1 an estimated 228 million (95% confidence interval [CI]: 206–258 million) persons worldwide have malaria, with 405,000 deaths in 2018. Eighty per cent of global malaria deaths happened in 14 sub-Saharan African countries and India.2 This mortality in sub-Saharan Africa is aggravated by the high proportion of deaths in under 5 years age groups (approximately 90% malaria-related deaths), more than any other malaria-endemic region.2 In general, pregnant women and children under the age of 5 years are the most vulnerable due to depressed immunity or the absence of partial immunity against the disease.3 Thus, strategies for reducing infection and disease burden in pregnant women, infants and children, groups bearing the highest burden of the disease, are increasingly urgent. Intermittent preventive treatment (IPT) is one such strategy. The World Health Organization (WHO) recommends IPT in pregnancy (IPTp) with sulfadoxine-pyrimethamine (SP) in all areas of Africa with moderate to high malaria transmission.4,5 In sub-Saharan Africa, 39 countries adopted IPTp between 1993 and 2007.5 In addition, the concept of IPT has been extended to include infants (IPTi) and children (IPTc; now referred to as SMC (seasonal malaria chemoprevention)).
The sub-Saharan African country Equatorial Guinea has a total population of 1.31 million (2018). According to WHO data, malaria causes 15% of all deaths in children under age 5 in Equatorial Guinea.4 Bioko, an island of Equatorial Guinea off the coast of Cameroon, has historically high malaria transmission.6 The most recent studies focused on detecting the prevalence of mutations in Pfmdr1 and Pfcrt on Bioko Island.7,8 SP has been used as a second-line therapy and IPTp on Bioko for several decades.8,9 Although IPT (IPTp, IPTi, IPTc) as a malaria control strategy has been shown to have a positive impact in preventing disease-related death in IPT-treated individuals, there are challenges due to the emergence of resistance to the drugs used for IPT treatment. From 1992 to 1999, the Carlos III Institute of Health (Madrid, Spain), working in collaboration with Equatorial Guinea’s Ministry of Health, conducted a study on Bioko Island and concluded that resistance to SP was approximately 25% among children under 5 years of age.10,11 In 2006, following the WHO recommendation to use combination therapy for malaria treatment, artemisinin-based combination therapy (ACT) was adopted as a first-line treatment,12,13 but SP is still used for IPT and in combination with artemisinin for the treatment of uncomplicated malaria, which may have led to Plasmodium falciparum isolates undergoing sustainable selection pressure.
Parasite resistance to SP is influenced by single-nucleotide polymorphisms (SNPs) of the P. falciparum dihydrofolate reductase (Pfdhfr) and dihydropteroate synthetase (Pfdhps) genes, which encode proteins involved in the folate biosynthesis pathway.14 Previous report revealed that the combination of Pfdhfr-Pfdhps mutations significantly correlated with the treatment failure.15 For the Pfdhfr gene, mutations N51I, C59R, S108N and I164L are associated with pyrimethamine resistance. Similarly, five mutations in Pfdhps (S436A, A437G, K540E, A581G and A613S/T) have been implicated in sulfadoxine resistance.16 Evidence suggests that SP resistance tends to increase as the result of the stepwise accumulation of SNPs in Pfdhps and Pfdhfr.6 Selective sweeps flanking Pfdhps and Pfdhfr have been shown in African populations as well.17–19 Under SP pressure, negative selection would begin to reduce the prevalence of the resistant genotypes, while recombination and mutation would begin to increase heterozygosity in markers flanking the resistance-conferring genes, degrading the selective sweep.19 All these observations underscore the need to monitor the distribution of parasite populations to mitigate the dispersal of resistant haplotypes in the region.
Unlike in other African countries, studies investigating Pfdhfr and Pfdhps mutations on Bioko Island are limited.10,12 Our recent study revealed a high prevalence of Pfdhfr and Pfdhps mutations associated with SP resistance among Bioko P. falciparum isolates.20 However, it remains unclear whether SP resistance has been decreasing or increasing in recent years since the Ministry of Health switched from SP to ACT for first-line treatment in 2006. Therefore, to ensure the prophylactic efficacy of IPT and to support the national antimalarial policy, the present study was carried out on Bioko Island at different time points to investigate the trend of molecular markers associated with SP resistance among P. falciparum isolates from Jan. 2011 to Dec. 2017. Additionally, further molecular evolutionary and population genetic approaches were applied to understand the evolution and spread of parasite drug resistance on Bioko Island.
Patients and Methods
Study Area
This study was carried out in the clinic of the Chinese medical aid team to the Republic of Equatorial Guinea, Malabo Regional Hospital of Bioko Island. Ethical approval was obtained from the Ethics Committee of Malabo Regional Hospital. Bioko is an island 32 km off the west coast of Africa and the northernmost part of Equatorial Guinea. The island has a population of 334,463 inhabitants (2015 census, of which approximately 90% live in Malabo, the capital city of Equatorial Guinea) and a humid tropical environment. Malaria due to P. falciparum is still the major public health issue on the island.21 The Bioko Island Malaria Control Project (BIMCP) has committed to reducing the burden of malaria on Bioko Island through methods such as concerted vector control, improved case management, and various educational interventions for the past 15 years. As a result of the vector control interventions and case management since 2004, the parasite prevalence on Bioko has decreased from over 45% in 2004 to 8.5% in 2016, and the entomological inoculation rate has decreased from more than 1000 before 2004 to 14 in 2015.22
Sample Collection
A total of 179 blood spot samples were collected from patients with uncomplicated malaria from January 2011 to December 2017. The included patients were aged between 4 months and 80 years and were residents of Bioko Island, collected in outpatient service of Malabo Regional Hospital with active case detection. All patients confirmed P. falciparum malaria cases identified by microscopic examination and an immuno-colloidal gold test kit (ICT Diagnostics). The patients were classified into uncomplicated malaria states according to WHO criteria, which were defined as positive smears for P. falciparum and the presence of fever (≥37.5°C).
Verbal informed consent was obtained from all participating subjects or their parents, and this study, as well as the consent process, was approved by the Ethics Committee of Malabo Regional Hospital. Dried blood spots were collected on day zero of enrolment through finger prick, with the blood spotted onto Whatman 903® filter paper (GE Healthcare, Pittsburgh, USA). Laboratory screening for malaria was performed using rapid diagnostic tests and confirmed using microscopic examination of blood smears. For quality control, archived malaria-positive microslides were re-examined, and parasite density was recorded. Plasmodium species were identified by real-time PCR followed by high-resolution melting.23 pGEM-T standard plasmids for four human Plasmodium species, including P. falciparum, P. ovale, P. malariae and P. vivax, which were kindly provided by Dr. Cao J (Jiangsu Institute of Parasitic Diseases, Wuxi, Jiangsu Province, China), were used as controls.
DNA Extraction and Sequencing
DNA was extracted from the dried blood spots following the Chelex-100 extraction procedure described in our previous report.7 Mutation screening for Pfdhfr and Pfdhps was performed by nested PCR as previously reported, with minor modifications.20 All primers were ordered from Genewiz (Soochow, China). The PCR products were separated by 2% agarose gel electrophoresis and stained with Gel Red (Genewiz, Soochow, China). The correct genes were identified based on size using an ultraviolet transilluminator. Next, the PCR fragments were purified and sequenced using corresponding primers by Sanger sequencing (Genewiz, Soochow, China). The sequences were analysed using Lasergene analysis software (DNA Star, Madison, WI) with 3D7 Pfdhfr and Pfdhps sequences as references (GenBank accession numbers: XM_001351443 and Z30654.1).
Data Analysis
Samples with multiple peaks at any genotyped SNP codon (mixed genotype) were excluded from SNP and haplotype analyses. The prevalence of mutations and haplotypes between collection years were compared using the chi-square or Fisher’s exact test, as appropriate (SPSS v.17 statistical software). Statistical analyses were performed at the 5% significance level and 95% CI. The inferred haplotype network was constructed by the median-joining network algorithm24 with the POPART program.25 Pfdhfr and Pfdhps sequences from 20 other countries (18 for Pfdhps and 18 for Pfdhfr) were downloaded from NCBI or our previous study26 to analyse global genetic diversity.27–36
Results
General Characteristics
From January 2011 to December 2017, we enrolled 179 patients with uncomplicated malaria from Bioko Island, Equatorial Guinea. Among the cases, 90.5% (162/179) and 77.1% (138/179) of the samples collected were successfully amplified and sequenced for the Pfdhfr and Pfdhps genes, respectively. These nucleotide sequences have been deposited in GenBank under accession numbers MN598219-MN598518. Multiclonal samples were excluded from this study. The median (interquartile range [IQR]) age of the patients was 39 years (14 to 68), and 36.9% of them were women. The median (IQR) parasite density was 4650 parasites/μL (340 to 63,120). Because the percentages are low, samples with mixed genotypes were not included when calculating the allele prevalence and reconstructing the various haplotypes (data not shown).
Frequency of Pfdhfr and Pfdhps Mutations
Pfdhfr and Pfdhps were examined to assess the frequency and variation trend of SP-resistance mutations in Bioko parasites from 2011 to 2017. The detailed information is shown in Tables 1 and 2. The three Pfdhfr mutations N51I, C59R and S108N were detected in 97.5%, 95.7% and 98.1% of the samples, respectively (Table 1). No other Pfdhfr mutations associated with SP resistance were observed. Five Pfdhps mutations associated with SP resistance were found. The mutation A437G was the most prevalent, being found in close to 90% of the samples (88.4%), and this frequency was significantly higher than the other mutations (p<0.05). Among the other four mutations, the frequency of S436A was relatively high (25.4%), followed by A613S (7.2%), K540E (5.1%) and A581G (1.4%) (Table 2). The frequency of most molecular markers in the Pfdhfr and Pfdhps genes exhibited a slowly increasing trend over the 7 years. However, there was no significant difference among the time periods (p>0.05).
 |
Table 1 The Frequency and Variation Trend of SP-Resistance Mutations in Pfdhfr Among Bioko Parasites from 2011 to 2017 |
 |
Table 2 The Frequency and Variation Trend of SP-Resistance Mutations in Pfdhps Among Bioko Parasites from 2011 to 2017 |
Frequency of Pfdhfr and Pfdhps Haplotypes
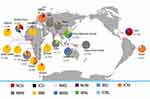
Four Pfdhfr and 10 Pfdhps haplotypes were obtained from Bioko parasites (Table S1). The frequency of the Pfdhfr triple-mutant haplotype (IRNI) was significantly higher (95.7%, 155/162) than was that of double-mutant haplotypes (ICNI, 2.5%; NRNI, 0.6%) and wild-type haplotypes (NCSI, 1.8%) (p<0.05). This haplotype profiling was consistent with neighbouring countries, including Gabon,37 Cameroon34 and Nigeria38 (Figure 1). IRNI and NRNI are the two most common Pfdhfr haplotypes worldwide. In the vast majority of Africa, IRNI is prevalent in many endemic regions, including Bioko Island, Gabon,37 Cameroon,33 Kenya36 and Malawi.32 NRNI also appears frequently, with a high percentage occurring in countries outside of Africa, such as Iran (92.3%),39 Nepal (90.6%),40 India (63.3%),34 Indonesia (62.5%),31 and Papua New Guinea (95.7%).30
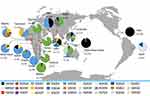
The frequency of the Pfdhps single-mutant haplotype (SGKAA) was predominant among the Bioko parasitic population (63.8%). Four haplotypes (SAKAS, SGKAS, SGKGS and AGKGS) were only found in one isolate. Interestingly, the Pfdhps haplotype diversity displays a slowly increasing trend since 2011 (Table S1). This haplotype profiling was consistent with neighbouring countries Angola35 and Gabon37 (Figure 2). A significant difference in haplotype profiles was found between East African countries and West African countries along the Atlantic Ocean (Figure 2). SGKAA has been reported to be the dominant haplotype in most African countries along the Atlantic Ocean, including Cameroon,33 Nigeria,38 Gabon,37 Angola,35 Democratic Republic of Congo41 and Bioko Island, and SGEAA is the major haplotype in East African countries, such as Uganda,29 Tanzania,29 Kenya36 and Malawi.32 The wild-type haplotype (SAKAA) is rare in almost all African and Asian countries but prevalent in Haiti42 and Papua New Guinea.30
The combination of 3 Pfdhfr and 5 Pfdhps haplotypes was found in 128 samples, and 14 Pfdhfr-Pfdhps haplotypes were determined; triple Pfdhfr and single Pfdhps mutations (IRNI-SGKAA) were most prevalent (59.4%, 76/128), followed by triple Pfdhfr and quintuple Pfdhps mutations (IRNI-AGKAA) (10.9%, 14/128) and triple Pfdhfr and single Pfdhps mutations (IRNI-AAKAA) (7.8%, 10/128). The wild-type haplotype (NCSI-SAKAA) was not found in this study (Table S2).
Relationship Among Pfdhfr and Pfdhps Sequences Between Bioko and Global Parasites
To assess the relationship among Pfdhfr sequences between Bioko and global P.falciparum isolates, 603 sequences, including 352 from this study and our previous study25 and 251 of other African and Asian countries (India, Congo, and Nigeria) downloaded from NCBI,34,38,41 were used to construct a haplotype network (Figure 3). For all SNPs (nonsynonymous and synonymous mutations) located in the Pfdhfr gene, 21 distinct haplotypes (H_1 to H_21) were found worldwide (Figure 3). H_1, the most prevalent haplotype in African countries, is associated with Asian sequences through H_3 and H_2. In addition, the network suggested that the majority of rare African haplotypes may have originated from the H_1 haplotype. Notably, approximately half of the Pfdhfr haplotypes only share sequences with Bioko parasites (10/21), indicating a high number of genetic polymorphisms of this gene in the Bioko parasitic population.
A total of 589 Pfdhps sequences were used to construct another haplotype network (Figure 4). These sequences included 327 from this study and our previous study25 and 262 of other African and Asian countries (India, Congo, and Nigeria) downloaded from NCBI.34,38,41 For all SNPs (nonsynonymous and synonymous mutations) in the Pfdhps gene, a total of 23 distinct haplotypes (H_1 to H_23) were identified (Figure 4). Eight haplotypes are shared by Pfdhps sequences from at least two countries; 15 haplotypes were found for one sequence. Consistent with Pfdhfr, most of them were found to be distributed on Bioko (8/15), which also indicates rich genetic polymorphisms among Bioko parasites. The most prevalent haplotype, H_1, with a percentage of 53.1%, has been found in Asia and 11 countries in Africa. Most haplotypes are shared with intercontinental Pfdhps sequences.
Discussion
In the current study, a large amount of mutations in the Pfdhfr and Pfdhps genes related to SP resistance was found among samples collected on Bioko Island at different time periods. Though there is still a lack of direct evidence to demonstrate that the treatment failures linked to SP resistance on Bioko Island, the frequencies of most mutations showed a slow and persistent increasing trend over the 7 years (2011–2017) investigated. When a country or region withdraws a given treatment due to drug resistance, the sensitive parasite population increases its presence with respect to the resistant population over a given time period. On Bioko, although the Ministry of Health withdrew SP as a first-line treatment for uncomplicated malaria after 2006,12 SP is still used as an alternative drug for IPTp and as one of the compositions of ACT therapy, which may explain the increasing trend of molecular markers associated with SP resistance in the region.
Similar to previous reports,12,20 two Pfdhfr mutations (N51I and S108N) in the Bioko parasite population kept its high frequency during the study period (2011–2017) and even reached 100% in recent years (2013–2017). This result might indicate that the two mutations have been fixed in the Bioko parasite population, which might influence the effectiveness of SP applied on Bioko because these mutations are related to pyrimethamine resistance. Pfdhfr I164L is an important mutation implicated in the effectiveness of SP as IPT, both in children under 5 years old and pregnant women,12 and this mutation has appeared on the Equatorial Guinea mainland. Fortunately, this mutation was absent among Bioko isolates.
The current WHO recommendations suggest that SP-IPTp should be discontinued if the frequency of K540E is more than 50.0% and that of A581G is above 10.0%. Fortunately, the frequencies of these two mutations on Bioko Island did not reach the standard yet. For this reason, SP-IPTp should still be effective on Bioko Island. Nonetheless, the Bioko Malaria Control Program (BIMCP) and the Ministry of Health and Social Welfare of Equatorial Guinea should implement measures on the island to control the spread of these mutations. Moreover, it is important to prevent an increase in the frequency of these two mutations to avoid reducing the efficiency of IPT-SP.
Consistent with our previous finding (2013–2014), this study revealed an intense level of Pfdhfr triple mutants (IRNI) and a single-mutant Pfdhps haplotype (SGKAA) among Bioko parasites.20 Global data27–36 were also applied in the comparison analysis, and the patterns for African countries in this study were in line with previous research.43 High similarity of the distribution of Pfdhfr and Pfdhps haplotypes was found between Bioko Island and its neighbouring countries. Combined with information from the network analysis, this result indicates that the origin of the Pfdhfr and Pfdhps haplotypes in Bioko parasites is most likely related to the African mainland.
SP-resistant parasites can be classified into three groups: “partially resistant”, “fully resistant” and “super resistant” based on Pfdhfr-Pfdhps haplotypes. In particular, the combination of the triple Pfdhfr mutant (N51I, C59R, S108N; haplotype IRNI) and Pfdhps A437G (haplotype SGKAA) confers partial resistance; the combination of the triple Pfdhfr mutant (N51I, C59R, S108N; haplotype IRNI) and double Pfdhps mutant (A437G, K540E; haplotype SGEAA) confers full resistance; and the combination of the triple Pfdhfr mutant (N51I, C59R, S108N; haplotype IRNI) and triple Pfdhps mutant (A437G, K540E, A581G; haplotype SGEGA) confers super resistance.44 These different combinations of mutations or haplotypes might affect the results of IPT in infected pregnant women and children. However, our data, fortunately, demonstrated that partially SP-resistant (IRNI-SGKAA) parasite was moderately popular and fully SP-resistant (IRNI-SGEAA) parasite was still rare on Bioko Island. Indeed, no super SP-resistant parasites were found on Bioko Island. Based on current evidence, IPTp-SP remains effective in preventing the adverse consequences of malaria on maternal and foetal outcomes on Bioko Island.
Conclusion
Historically, SP has been extensively used on Bioko by the Ministry of Health of Equatorial Guinea. Our findings reveal the general existence of SP-resistance markers on Bioko Island even after the replacement of SP for first-line treatment of uncomplicated malaria in 2006. Although no significant difference was observed in different time periods (p>0.05), our study found a slow increasing trend of the frequencies of these SP-resistance markers among Bioko parasites over the 7 years evaluated. Therefore, it is very important to control the use of SP and to ensure that it is used exclusively for IPT in children under 5 years old and pregnant women. Furthermore, to halt the SP mutations, it is urgent to call for careful monitoring of genotypic resistance markers and in vivo validation of IPT efficacy. Nowadays, SP is not recommended for use as a daily treatment on Bioko Island, either alone or in combination with another treatment (artesunate+SP; AS+SP). This strategy will limit increases in the number of mutations, the occurrence of new mutations, and in particular, the spread of mutations to protect the population more effectively. In addition, continuous molecular monitoring and additional control efforts are urgently needed.
Abbreviations
IPT, intermittent preventive treatment; IPTp, intermittent preventive treatment in pregnancy; SP, sulfadoxine-pyrimethamine; ACT, artemisinin-based combination therapy; SNP, single-nucleotide polymorphisms; P.falciparum, Plasmodium falciparum; P.ovale, Plasmodium ovale; P.malariae, Plasmodium malariae; P.vivax, Plasmodium vivax; Pfdhfr, Plasmodium falciparum dihydrofolate reductase; Pfdhps, Plasmodium falciparum dihydropteroate synthase.
Acknowledgments
The authors thank the Department of Health of Guangdong Province and the Department of Aid to Foreign Countries of Ministry of Commerce of the People’s Republic of China for their help. The authors also thank Santiago-m Monte-Nguba for his technical help during the sample collection and diagnosis.
Funding
This work was partially supported by the Natural Science Foundation of Guangdong Province (Grant No. 2016A03031311 to Jiang-Tao Chen; 2018A030307074 to Yu-Zhong Zheng), the Guangdong Science and Technology Project (Grant No. 2016A030303064 to Guang-Cai Zha), the Foundation for Innovative Research Team of Hubei University of Medicine (Grant No. FDFR201603 to Jian Li), the National Natural Science Foundation of China (Grant No. 81802046 to Jian Li), 2020 Chaozhou’s First Batch of Special Science and Technology Plans, and 2019 Guangdong Province Key Discipline Research Project.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. World malaria report; 2019. Available from: https://www.who.int/news-room/feature-stories/detail/world-malaria-report-2019.
2. Grais RF, Laminou IM, Woi-Messe L, et al. Molecular markers of resistance to amodiaquine plus sulfadoxine-pyrimethamine in an area with seasonal malaria chemoprevention in south central Niger. Malar J. 2018;17(1):98. doi:10.1186/s12936-018-2242-4
3. Ruizendaal E, Tahita MC, Geskus RB, et al. Increase in the prevalence of mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates collected from early to late pregnancy in Nanoro, Burkina Faso. Malar J. 2017;16(1):179. doi:10.1186/s12936-017-1831-y
4. Guerra M, de Sousa B, Ndong-Mabale N, Berzosa P, Arez AP. Malaria determining risk factors at the household level in two rural villages of mainland Equatorial Guinea. Malar J. 2018;17(1):203. doi:10.1186/s12936-018-2354-x
5. Tessema SK, Kassa M, Kebede A, et al. Declining trend of Plasmodium falciparum dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant alleles after the withdrawal of sulfadoxine-pyrimethamine in North Western Ethiopia. PLoS One. 2015;10(10):e0126943. doi:10.1371/journal.pone.0126943
6. Nkoli Mandoko P, Rouvier F, Matendo Kakina L, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine-pyrimethamine in the Democratic Republic of the Congo: emergence of highly resistant Pfdhfr/Pfdhps alleles. J Antimicrob Chemother. 2018;73(10):2704–2715. doi:10.1093/jac/dky258
7. Li J, Chen J, Xie D, et al. Molecular mutation profile of Pfcrt and Pfmdr1 in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Infect Genet Evol. 2015;36:552–556. doi:10.1016/j.meegid.2015.08.039
8. Li J, Chen J, Xie D, et al. High prevalence of Pfmdr1 N86Y and Y184F mutations in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Pathog Glob Health. 2014;108(7):339–343. doi:10.1179/2047773214Y.0000000158
9. Guerra M, Neres R, Salgueiro P, et al. Plasmodium falciparum Genetic Diversity in Continental Equatorial Guinea before and after Introduction of Artemisinin-Based Combination Therapy. Antimicrob Agents Chemother. 2017;61(1).
10. Benito A, Roche J, Molina R, Amela C, Alvar J. In vitro susceptibility of Plasmodium falciparum to chloroquine, amodiaquine, quinine, mefloquine, and sulfadoxine-pyrimethamine in Equatorial Guinea. Am J Trop Med Hyg. 1995;53(5):526–531. doi:10.4269/ajtmh.1995.53.526
11. Charle P, Berzosa P, de Lucio A, Raso J, Nseng Nchama G, Benito A. Artesunate/amodiaquine malaria treatment for Equatorial Guinea (Central Africa). Am J Trop Med Hyg. 2013;88(6):1087–1092. doi:10.4269/ajtmh.12-0290
12. Berzosa P, Esteban-Cantos A, Garcia L, et al. Profile of molecular mutations in Pfdhfr, Pfdhps, Pfmdr1, and Pfcrt genes of Plasmodium falciparum related to resistance to different anti-malarial drugs in the Bata District (Equatorial Guinea). Malar J. 2017;16(1):28. doi:10.1186/s12936-016-1672-0
13. Manore CA, Teboh-Ewungkem MI, Prosper O, Peace A, Gurski K, Feng Z. Intermittent Preventive Treatment (IPT): its role in averting disease-induced mortality in children and in promoting the spread of antimalarial drug resistance. Bull Math Biol. 2019;81(1):193–234. doi:10.1007/s11538-018-0524-1
14. Spalding MD, Eyase FL, Akala HM, et al. Increased prevalence of the Pfdhfr/phdhps quintuple mutant and rapid emergence of Pfdhps resistance mutations at codons 581 and 613 in Kisumu, Kenya. Malar J. 2010;9(1):338. doi:10.1186/1475-2875-9-338
15. Mohamed AO, Hussien M, Mohamed A, et al. Assessment of Plasmodium falciparum drug resistance molecular markers from the Blue Nile State, Southeast Sudan. Malar J. 2020;19(1):78. doi:10.1186/s12936-020-03165-0
16. Osarfo J, Tagbor H, Magnussen P, Alifrangis M. Molecular markers of Plasmodium falciparum drug resistance in parasitemic pregnant women in the middle forest belt of Ghana. Am J Trop Med Hyg. 2018;98(6):1714–1717. doi:10.4269/ajtmh.18-0009
17. McCollum AM, Schneider KA, Griffing SM, et al. Differences in selective pressure on dhps and dhfr drug resistant mutations in western Kenya. Malar J. 2012;11(1):77. doi:10.1186/1475-2875-11-77
18. Artimovich E, Kapito-Tembo A, Pensulo P, et al. The effect of local variation in malaria transmission on the prevalence of sulfadoxine-pyrimethamine resistant haplotypes and selective sweep characteristics in Malawi. Malar J. 2015;14(1):387. doi:10.1186/s12936-015-0860-7
19. Artimovich E, Schneider K, Taylor TE, et al. Persistence of sulfadoxine-pyrimethamine resistance despite reduction of drug pressure in Malawi. J Infect Dis. 2015;212(5):694–701. doi:10.1093/infdis/jiv078
20. Jiang T, Chen J, Fu H, et al. High prevalence of Pfdhfr-Pfdhps quadruple mutations associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Malar J. 2019;18(1):101. doi:10.1186/s12936-019-2734-x
21. Cook J, Hergott D, Phiri W, et al. Trends in parasite prevalence following 13 years of malaria interventions on Bioko island, Equatorial Guinea: 2004–2016. Malar J. 2018;17(1):62. doi:10.1186/s12936-018-2213-9
22. Malaria Control and Elimination. Available from: https://www.mcdinternational.org/bimcp.
23. Wang SQ, Zhou HY, Li Z, et al. Quantitative detection and species identification of human Plasmodium spp. by using SYBR Green I based real-time PCR. Zhong Guo Xue Xi Chong Bing Fang Zhi Za Zhi. 2011;23(6):677–681.
24. Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16(1):37–48. doi:10.1093/oxfordjournals.molbev.a026036
25. Leigh JW, Bryant D, Nakagawa S, Nakagawa S. Popart: full-feature software for haplotype network construction. Methods Ecol Evol. 2015;6(9):1110–1116. doi:10.1111/mee3.2015.6.issue-9
26. Yao Y, Wu K, Xu M, et al. Surveillance of genetic variations associated with antimalarial resistance of Plasmodium falciparum isolates from returned migrant workers in Wuhan, Central China. Antimicrob Agents Chemother. 2018;62(9). doi:10.1128/AAC.02387-17.
27. Bai Y, Zhang J, Geng J, et al. Longitudinal surveillance of drug resistance in Plasmodium falciparum isolates from the China-Myanmar border reveals persistent circulation of multidrug resistant parasites. Int J Parasitol Drugs Drug Resist. 2018;8(2):320–328. doi:10.1016/j.ijpddr.2018.05.003
28. Bamaga OA, Mahdy MA, Lim YA. Frequencies distribution of dihydrofolate reductase and dihydropteroate synthetase mutant alleles associated with sulfadoxine-pyrimethamine resistance in Plasmodium falciparum population from Hadhramout Governorate, Yemen. Malar J. 2015;14(1):516. doi:10.1186/s12936-015-1035-2
29. Baraka V, Delgado-Ratto C, Nag S, et al. Different origin and dispersal of sulfadoxine-resistant Plasmodium falciparum haplotypes between Eastern Africa and Democratic Republic of Congo. Int J Antimicrob Agents. 2017;49(4):456–464. doi:10.1016/j.ijantimicag.2016.12.007
30. Barnadas C, Timinao L, Javati S, et al. Significant geographical differences in prevalence of mutations associated with Plasmodium falciparum and Plasmodium vivax drug resistance in two regions from Papua New Guinea. Malar J. 2015;14(1):399. doi:10.1186/s12936-015-0879-9
31. Basuki S, Fitriah RS, Budiono DYP, Uemura H. Two novel mutations of Pfdhps K540T and I588F, affecting sulphadoxine-pyrimethamine-resistant response in uncomplicated falciparum malaria at Banjar district, South Kalimantan Province, Indonesia. Malar J. 2014;13(1):135. doi:10.1186/1475-2875-13-135
32. Bwijo B, Kaneko A, Takechi M, et al. High prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003;85(3):363–373. doi:10.1016/S0001-706X(02)00264-4
33. Chauvin P, Menard S, Iriart X, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine-pyrimethamine in pregnant women in Yaounde, Cameroon: emergence of highly resistant Pfdhfr/Pfdhps alleles. J Antimicrob Chemother. 2015;70(9):2566–2571. doi:10.1093/jac/dkv160
34. Das MK, Chetry S, Kalita MC, Dutta P. Evidence of triple mutant Pfdhps ISGNGA haplotype in Plasmodium falciparum isolates from North-east India: an analysis of sulfadoxine resistant haplotype selection. Genom Data. 2016;10:144–150. doi:10.1016/j.gdata.2016.11.001
35. Kaingona-Daniel EP, Gomes LR, Gama BE, et al. Low-grade sulfadoxine-pyrimethamine resistance in Plasmodium falciparum parasites from Lubango, Angola. Malar J. 2016;15(1):309. doi:10.1186/s12936-016-1358-7
36. Lucchi NW, Okoth SA, Komino F, et al. Increasing prevalence of a novel triple-mutant dihydropteroate synthase genotype in Plasmodium falciparum in western Kenya. Antimicrob Agents Chemother. 2015;59(7):3995–4002. doi:10.1128/AAC.04961-14
37. Voumbo-Matoumona DF, Kouna LC, Madamet M, Maghendji-Nzondo S, Pradines B, Lekana-Douki JB. Prevalence of Plasmodium falciparum antimalarial drug resistance genes in Southeastern Gabon from 2011 to 2014. Infect Drug Resist. 2018;11:1329–1338. doi:10.2147/IDR.S160164
38. Oboh MA, Singh US, Antony HA, et al. Molecular epidemiology and evolution of drug-resistant genes in the malaria parasite Plasmodium falciparum in southwestern Nigeria. Infect Genet Evol. 2018;66:222–228. doi:10.1016/j.meegid.2018.10.007
39. Rouhani M, Zakeri S, Pirahmadi S, Raeisi A, Djadid ND. High prevalence of Pfdhfr-Pfdhps triple mutations associated with anti-malarial drugs resistance in Plasmodium falciparum isolates seven years after the adoption of sulfadoxine-pyrimethamine in combination with artesunate as first-line treatment in Iran. Infect Genet Evol. 2015;31:183–189. doi:10.1016/j.meegid.2015.01.020
40. Ranjitkar S, Schousboe ML, Thomsen TT, et al. Prevalence of molecular markers of anti-malarial drug resistance in Plasmodium vivax and Plasmodium falciparum in two districts of Nepal. Malar J. 2011;10(1):75. doi:10.1186/1475-2875-10-75
41. Ruh E, Bateko JP, Imir T, Taylan-Ozkan A. Molecular identification of sulfadoxine-pyrimethamine resistance in malaria infected women who received intermittent preventive treatment in the Democratic Republic of Congo. Malar J. 2018;17(1):17. doi:10.1186/s12936-017-2160-x
42. Morton LC, Huber C, Okoth SA, et al. Plasmodium falciparum drug-resistant haplotypes and population structure in postearthquake Haiti, 2010. Am J Trop Med Hyg. 2016;95(4):811–816. doi:10.4269/ajtmh.16-0214
43. Okell LC, Griffin JT, Roper C. Mapping sulphadoxine-pyrimethamine-resistant Plasmodium falciparum malaria in infected humans and in parasite populations in Africa. Sci Rep. 2017;7(1):7389. doi:10.1038/s41598-017-06708-9
44. Naidoo I, Roper C. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol. 2013;29(10):505–515. doi:10.1016/j.pt.2013.08.002
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.