Back to Journals » Cancer Management and Research » Volume 13
Transcriptome Signature of Immune Cells Post Reovirus Treatment in KRAS Mutated Colorectal Cancer
Authors Fogel EJ, Samouha A, Goel S, Maitra R
Received 12 June 2021
Accepted for publication 6 August 2021
Published 27 August 2021 Volume 2021:13 Pages 6743—6754
DOI https://doi.org/10.2147/CMAR.S324203
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Elisha J Fogel,1 Avishai Samouha,1 Sanjay Goel,2 Radhashree Maitra1,2
1Department of Biology, Yeshiva University, New York, NY, 10033, USA; 2Albert Einstein College of Medicine at Montefiore Medical Center, Bronx, NY, 10461, USA
Correspondence: Radhashree Maitra
Department of Biology, Yeshiva University, 500 W 185th Street, New York, NY, 10033, USA
Tel +1 718-801-1359
Email [email protected]
Purpose: Reovirus propagates with high efficiency in KRAS mutated colorectal cancer (CRC). About 45– 50% of CRC patients possess a KRAS mutation. Oncolytic reovirus treatment in combination with chemotherapy was tested in patients possessing KRAS mutated metastatic CRC. This study evaluates the biological responses to reovirus treatment by determining the gene expression patterns in RAS-related signaling pathways.
Methods: Reovirus was administered as a 60-min intravenous infusion for 5 consecutive days every 28 days, at a tissue culture infective dose (TCID50) of 3× 1010. Peripheral blood mononuclear cells (PBMCs) were isolated from whole-blood pre- and post-reovirus administration at 48 hr, day-8, and day-15. Clariom_D_Human_Assay was used to determine the expression of vital genes compared to pre-reovirus treatment by RNA sequencing. Using exported sample signals, ΔΔCt method was used to analyze the fold changes of genes within seven gene pathways. Significance was calculated by students-two-tail-t-test. Hierarchical clustering dendrogram was constructed by calculating Pearson’s correlation coefficients.
Results: As compared to the control, SOS1[48 hr; 2.49X], RRAS [48 hr; 2.24X], PIK3CB [D8, D15; 2.27X, 3.16X], MIR 16– 2 [D15; 1.70X], CHORDC1 [48 hr, D15; 1.89X, 4.54X], RTN4 [48 hr; 4.66X], FAM96A [48 hr; 4.54X], NFKB [D8, D15; 19.0X, 1.42X], CASP8 [D8, D15; 2.11X, 1.77X], and CASP9 [D8; 1.45X] are upregulated post-reovirus. NOS3 [D15; 0.61X], SYNE1 [D8, D15; 0.78X, 0.71X], ANGPT1 [D8; 0.62X], VEGFB [48 hr, D8, D15; 0.44X, 0.28X, 0.28X], JUN [D15; 0.69X], and IGF2 [D8; 0.73X] are downregulated post-reovirus. Fold change values were significant [p< 0.05].
Conclusion: This study highlights reovirus as a novel treatment option for KRAS mutated CRC and showcases its effect on the expression of crucial genes.
Keywords: transcriptome, reovirus, CRC, KRAS, CASP8, CHORDC1, RTN4, VEGFB
Introduction
RNA molecules play significant roles in biological functions, including, genetic coding/decoding, gene regulation and gene expressions.1 The identification of key RNA sequences in proteins and genes, as well as the understanding of the roles they play, provides an overwhelming amount of knowledge on specific biological pathways.2,3 This can allow for manipulation of the expression and regulation of certain genes in beneficial pharmaceutical and treatment methods. RNA research made a breakthrough in early 2000s with the advent of next-generation RNA-Seq.1 Two noteworthy studies, which were some of the first to use RNA-Seq, were Nagalakshmi et al4 on the landscape of the yeast genome and Bainbridge et al on prostate cancer cell lines.5 The two methods by which researchers use to assemble transcriptomes are De novo and Genome guided. The de novo method does not require a known reference genome in order to reconstruct the transcriptome; rather, sequences are assembled as contigs, a set of overlapping DNA segments that, as a collective, embody a consensus region of DNA.6 In the genome guided method, RNA sequences are aligned within a specific, known, genome, similar to that in the DNA alignment process.7 In this study RNA-Seq was conducted on peripheral blood mononuclear cells (PBMC) obtained from a set of patients who underwent reovirus treatment for metastatic colorectal cancer.
The use of reovirus as a therapy for cancer is an up-and-coming intervention in the field of cancer biology.8–10 Reovirus, a proprietary version of unmodified human reovirus, is an oncolytic virus that can lyse cancer cells.11,12 This virus is being developed as an immune-oncological viral agent to treat hematological malignancies and various forms of neoplasms.13 Reovirus is a stable form of nonenveloped double-stranded RNA. In humans, infection usually results in an asymptomatic or mild illness, manifested with flu-like symptoms, including mild gastroenteritis.14 Given the virus’ oncolytic properties and its potential to reproduce well in cancer cells, interest in therapeutic reovirus treatment has been proposed since 1970’s.15 Reovirus has become a prolific experimental model for viral pathogenesis.
Reovirus has been shown to replicate remarkably well in cells that have a mutated KRAS gene.12 After infection within KRAS-mutated tumor cells, viral replication occurs resulting in cell death, which leads to viral progeny infecting nearby tumor cells. It is hypothesized that upstream activation of KRAS proteins may play a role in more than two-thirds of human metastatic cancers.16 This provides further evidence of the therapeutic potential of reovirus, specifically in KRAS-mutated tumors, and its potential as an alternative treatment method for metastatic colorectal cancer. This study utilized the Genome guided method of RNA-Seq in order to analyze seven different biological pathways and how they were affected by reovirus treatment in samples from KRAS mutated colorectal cancer.
Methods
Patient Selection
Blood samples were obtained from eight patients, all having KRAS-mutated mCRC. Five (5) patients had received reovirus as part of a Phase 1 clinical trial (NCT01274624). Three (3) patients were not enrolled in the trial but did receive equivalent background chemotherapy (i.e. FOLFIRI and bevacizumab).
Ethical Considerations
Ethics approval and consent to participate: The entire study was performed in compliance with Institutional and Federal guidelines for clinical research. The study was approved by the ethics committee of Montefiore Medical Center/Albert Einstein School of Medicine. All patient tissue and blood samples were drawn following written informed consent based on a local IRB approved consent form. This trial was conducted in accordance with the Declaration of Helsinki.
Institutional Review Board/Ethics Committee Approval and Informed Consent
All patient blood samples were drawn with informed consent based on an Albert Einstein College of Medicine IRB committee approved consent form (IRB# 07-10-376). Following the institutional review board’s (ethics committee) approval, blood samples were collected from eight patients with KRAS-mutated metastatic colorectal cancer at 0 (pre) and 48 hr, and on days 8 and 15. Peripheral blood mononuclear cells (PBMCs) were isolated for transcriptome analysis from eight patients. Five patients had received reovirus (pelareorep) along with FDA approved second-line chemotherapy (i.e. FOLFIRI and bevacizumab) as part of a Phase I clinical trial (NCT01274624), while three patients were not enrolled in this trial but did receive equivalent chemotherapy.
Data Sharing
The authors have shared individual deidentified participant data in the public domain to be available indefinitely. These raw data include peripheral mononuclear cell (PBMC) samples at four timepoints (pre-treatment, 48 hr, day 8 and day 15) for each individual involved in the clinical trial. This data can be accessed through the NCBI GEO Accession Viewer (Series GSE173636) using the following URL: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE173636
Reovirus Administration
Reovirus was supplied by Oncolytics Biotech, Inc. as a translucent to clear, colorless to light blue liquid in vials containing 7.2×1010 tissue culture infective dose (TCID50) per mL of reovirus in a phosphate-buffered solution and stored at minus 70°C. Reovirus was administered as a 60-min infusion for 5 consecutive days every 28 days, at a tissue culture infective dose (TCID50) of 3 × 1010/day. Plasma was collected pre-reovirus [0 hr], at 48 hr, Day 8 (D8) and Day 15 (D15) after first dose of reovirus.
Transcriptome Analysis
Transcriptome Analysis Console (TAC) Software: Thermo Fisher Scientific is a software, which conducts RNA-Seq via the Genome guided method. The TAC 4.0 uses Expression Console™ software (EC), which utilizes cell intensity files (*. CEL) to create probe-level summarization files. Thus, allowing for initial data quality examination of Affymetrix expression arrays.
Blood was collected into CPT tubes (BD Biosciences, Franklin Lakes, NJ) for isolation of PBMC (Peripheral Blood Mononuclear Cells). Total RNA was isolated from the patient's PBMC and reverse transcribed using a reverse transcription priming method from an engineered set of primers that exclude sequences that match ribosomal RNA (rRNA). These primers specifically prime non-ribosomal RNA from the sample, including both poly(A) and non-poly(A) mRNA and convert it into double-stranded cDNA using first- and second-strand enzyme from Kit. The templates are used for in vitro transcription reaction at 37°C for 16 hr to yield cRNA. The ss-cDNA generated from cRNA was chemically fragmented, biotin-labeled and made into a hybridization cocktail using hybridization kit (Catalog Number 900454) according to the Affymetrix Genechip protocol, which was then hybridized to Clariom D Gene Chip human probe array. The array image is generated by a high-resolution Gene Array Scanner 3000 7G (Thermo Fisher Scientific, Santa Clara, CA) and the Cq value for each gene expression of a patient at a given timepoint was recorded. Individual sample signals for each patient at each timepoint were extracted from the TAC 4.0 software, organized and compiled in Microsoft Office Excel. Gene expression data were analyzed by the 2−ΔΔCT method17 and normalized to “Pre” treatment (Day 0) expression. Two tailed t-test was used to determine statistical significance (p<0.05). Statistics were calculated using Microsoft Office Excel.
After the results were collected from the patients, the data was uploaded to Transcriptome Analysis Console 4.0.1 (TAC). Seven growth and apoptotic related pathways were observed as detailed in Table 1.
 |
Table 1 Pathway Sources |
Two thousand eighty-three genes, across all seven pathways, were filtered via the software’s filter function and their sample signals were exported to their respective Excel spreadsheets. This was done for each set of genes of the seven biological pathways which were studied. Sample signal data was exported for each patient timepoint (Pre, 48 hr, Day 8 and Day 15). Four Excel sheets, one for each timepoint, were created with the sample signals for all experimental patients. Fold changes were calculated. This was accomplished by comparing the 48 hr, Day 8 and Day 15 timepoints to the Pre timepoint, separately. First, the ΔΔCts (Difference in Cq values) were calculated for each timepoint; then, the 2−ΔΔCt (Actual fold change) was calculated using the data attained from the ΔΔCt results. The 2−ΔΔCt results that were obtained for each patient at that respected timepoint for every gene used to calculate the mean fold change. Upon obtaining the results from these calculations, all 2083 genes (many of which overlapped into different pathways) were filtered by a p-value of 0.05. This yielded 155 significant genes, across all pathways and timepoints. Of the 155 genes, 16 (Table 2) of them were chosen to be further analyzed by virtue of their levels of expression and their effects on KRAS mutated CRC proliferation.
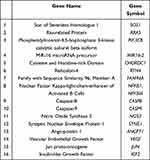 |
Table 2 Genes Analyzed |
Dendrogram Generation
Fold change values at all timepoints for the 16 genes analyzed were fed into XLSTAT as an agglomerative hierarchical clustering. The similarities of expression between the genes were quantified using the Pearson’s Correlation Coefficient.
Results
NF-κB, SOS1, and RRAS Gene Expression Indicated Successful Immune Response to Reovirus
SOS1 and RRAS were upregulated at 48 hr; NF-κB gene expression showed exceptional increase at Day 8 and a significant decrease at Day 15.
In PBMC samples of patients who underwent reovirus treatment, a statistically significant (p<0.05) increase in expression of SOS1 and RRAS was observed at 48 hr with fold changes of 2.49 [p=0.00339; SEM=0.706] and 2.24 [p=0.0186; SEM=0.340], respectively; both of these genes lost significance at D8 and D15. It was also observed that NF-κB had a fold change of 19.0 [p=0.050; SEM=6.34] at D8 and a fold change of 1.42 [p=0.0285; SEM=0.181] at D15. The major increase of NF-κB, followed by a drastic decrease at D15, implies that reovirus was effectively administered, thus promoting an immune response in the patients’ innate and adaptive immune systems. The loss of significance for SOS1 and RRAS post-48 hr, in conjunction with the major increase of NF-κB expression at D8, indicates that the major effects of reovirus treatment took effect at around D8. Consequently, this timepoint is where we find many of our genes starting to respond to reovirus treatment. These data are presented in Figure 1 and Supplementary Table 1.
 |
Figure 1 (A) The expression of SOS1 significant at 48 hr (B) The expression of RRAS significant at 48 hr (C) The expression of NFKB significant at D8 and D15. |
Genes Linked to Cell Proliferation Decrease in Transcription
Angiogenesis Related Genes, ANGPT1, VEGFB, and SYNE1 are Found to Be Significantly Downregulated
We observed a decrease in gene expression of ANGTP1 and VEGFB which were signified by a reduction in mean fold change values relative to Pre. This data is highlighted in Figure 2. ANGPT1 was found to be downregulated at D8 with a fold change of 0.62 [p=0.0120; SEM=0.113]. VEGFB was found to be downregulated at 48 hr with a fold change of 0.44 [p=0.00872; SEM=0.0882], at D8 with a fold change of 0.28 [p=0.00380; SEM=0.0410], and at D15 with a fold change of 0.28 [p=1.31×10−5; SEM=0.0456]. SYNE1 was found to be downregulated at D8 with a fold change of 0.78 [p=0.0392; SEM=0.0410] and at D15 with a fold change of 0.71 [p=0.0100; SEM=0.0585]. Down regulation indicates reduction in cell proliferation as well as decrease in angiogenesis. This data is highlighted in Figure 2.
 |
Figure 2 (A) The expression of VEGFB significant at all timepoints (B) The expression of ANGPT1 significant at D8 (C) The expression of SYNE1 significant at D8 and D15. |
Apoptotic Related Genes Increase in Reovirus Treated KRAS Mutated CRC
Key Apoptotic Genes CASP8 and CASP9 are Found to Be Upregulated; JUN and IGF2 are Found to Be Downregulated
CASP8 and CASP9 were upregulated at D8 with fold changes of 2.11 [p=0.0410; SEM=0.487] and 1.45 [p=0.00299; SEM= 0.105], respectively. In addition, CASP8 was upregulated at D15 with a fold change of 1.77 [p=0.0425; SEM=0.344]. These findings indicate a significant increase in apoptotic activity in KRAS mutated CRC post-reovirus treatment. JUN was found to be downregulated at D15 with a fold change of 0.69 [p=0.0358; SEM=0.110]. IGF2 was downregulated at D8 with a fold change of 0.73 [p=0.00727; SEM=0.0560]. Down regulation indicates a reduction in cell proliferation and an increase in cellular apoptosis. This data is highlighted in Figure 3.
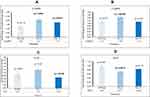 |
Figure 3 (A) The expression of CASP8 significant at D8 and D15 (B) The expression of CASP9 significant at D8 (C) The expression of JUN significant at D15 (D) The expression of IGF2 significant at D8. |
Genes Pertaining to miR in Lymphocytes Exhibit Significantly Positive Alteration Post Reovirus Treatment
RTN4 and FAM96A Were Upregulated at 48hr; CHORDC1 Was Upregulated at 48hr and D15; MIR16-2 Was Upregulated at D15
When compared to Pre mean Cq values, patients who underwent reovirus treatment exhibited higher levels of RTN4 and CHORDC1 gene expression. At 48 hr, RTN4, FAM96A, and CHORDC1 were upregulated with fold changes of 4.66 [p=0.00531; SEM=1.27], 4.54 [p=0.0364; SEM=1.27], and 1.89 [p=0.0352; SEM=0.369], respectively. Additionally, CHORDC1 and MIR16-2 were upregulated at D15 with fold changes of 4.54 [p=0.0254; SEM=1.51] and 1.70 [p=0.0204; SEM=0.301], respectively. These results showcase that in KRAS mutated CRC, post-reovirus treatment, patients possessed a significant increase in lymphocytic activity and strengthening of their adaptive immune systems. This data is highlighted in Figure 4.
Post Reovirus Treatment, Genes Within the RAS-Signaling Pathways of RAS, MAPK, EGFR, and PI3K-Akt, Showcased Gene Alteration
PIK3CB Was Upregulated at D8 and D15; NOS3 Was Downregulated at D15
Two significant RAS-signaling genes were identified across the four RAS-related gene signaling pathways. Upon comparison with mean Pre Cq values, CRC patients exhibited higher levels of PIK3CB gene expression, as well as lower levels of NOS3 gene expression post-reovirus treatment. At D8 PIK3CB was upregulated with fold a change of 2.27 [p=0.0000390; SEM=0.204]. At D15 PIK3CB was upregulated and NOS3 was downregulated with fold changes of 3.16 [p=0.00719; SEM= 0.846] and 0.61 [p=0.00889; SEM=0.107], respectively. These results confirmed that patients who underwent reovirus treatment saw tumor-suppressor effects of PIK3CB and NOS3. This data is highlighted in Figure 5 and Supplementary Table 1.
Pathway Correlations
Wanting to identify statistical correlations, four types of graphical interpretation of the data were created to interconnect the genes which were focused on. First, a Venn diagram was generated, showing common genes among the different pathways. A STRING interaction network was created, depicting the relationship between the 16 main genes. The third statistical representation was a heat map, which was generated using the mean fold changes from each timepoint (significant and not significant). Finally, a dendrogram-statistical analysis for the similarity of genes was created, with assessment based on the Pearson’s Correlation Coefficient. This data is displayed in Figure 6.
Discussion
Our research focused on analyzing the affects reovirus treatment had on KRAS mutated CRC on seven crucial biological pathways. We have gathered that across these seven pathways, apoptosis was induced, angiogenesis was inhibited, lymphocytic maturation and activity increased, and several RAS-related signaling genes were altered to aid the immune response to CRC. Reovirus induces alterations in several gene transcripts that can be correlated with oncolysis. Upregulation of apoptotic genes and downregulation of angiogenesis-related genes indicate that reovirus can be an effective CRC treatment in patients possessing KRAS mutated cells. The results clearly show the effect of reovirus. Patients receiving background chemotherapy but not the reovirus do not have any significant alterations in these genes at 48 hr, D8, or D15 [Supplementary Tables 1 and 2]. In this study, seven biological pathways were completely analyzed and assessed based on the efficacy of reovirus-altered gene expression on genes within these pathways.
The NF-κB family deals with controlling genes that regulate cellular differentiation and the transcription of cytokines and antimicrobial effectors.18 However, an additional characteristic of NF-κB is its contribution to development of immune cells and their survivability and its use as a transcription factor that promotes transcription of many genes including MAPK. Our data showed a drastic increase in gene expression of NF-κB at the D8 timepoint, suggesting a trigger in immune response due to the administration of reovirus.
This observed increase was enabled by the activity of SOS1, which encodes a protein that is a guanine nucleotide exchange factor for RAS proteins.19 By participating in RAS-related signal transduction pathways, SOS1 has been shown to increase NFκB activation by constitutive stimulation of the EGF receptor and phosphorylation of MAPK3.20 Our data show that SOS1 was overexpressed at 48 hr, a requirement for the EGF-dependent activation of NFκB that was witnessed significantly at D8. Additionally, this increase was followed by a fold change decrease to basal levels at the D15 timepoint, which signified a complete cycle of immune response to the treatment.
Angiogenesis is a biological process, which promotes the creation of blood vessels; observations of genes in the angiogenesis pathway can reveal the state of cancer cell progression or regression.21 ANGPT1 directly plays a role in the creation of blood vessels and their maturation as well as vascular development.22 In metastatic CRC, this gene is generally upregulated, pointing cellular proliferation of cancer cells; however, in our data we see a decrease of ANGTP1 at the D8 timepoint. While this gene loses significance at D15, another key angiogenesis-related gene, VEGFB, was observed to be significant and was down regulated at all timepoints. SYNE1, a gene that helps encode a spectrin repeat containing protein, which is expressed in peripheral blood lymphocytes,23 was found to be downregulated at both D8 and D15. The downregulation of VEGFB at every timepoint, in conjunction with the downregulation of ANGPT1 and SYNE1, proves the decrease in cancer cell proliferation and angiogenesis in CRC.
Apoptosis, a form of programmed cell death, is controlled by aspartate-specific cysteine proteases (caspases) and functions in maintaining homeostasis by the removal of damaged cells. We have identified multiple genes, which function in the apoptotic pathway, that have been affected via reovirus treatment. The CASP8 and CASP9 are two important members of the pathway that are significantly upregulated favoring apoptosis. CASP8 not only regulates apoptosis but has also been found to stimulate cell proliferation, malignant transformation and tumor progression when it is dysfunctional or has significantly low gene expression.24 Thus, these genes play a crucial role in tumor suppression.
JUN is an intron-less proto-oncogene, which has a key role in cellular proliferation and apoptosis.25 When overexpressed, JUN has been seen to suppress cellular apoptosis. This function has shown to be highly exacerbated by exposure to UV radiation.26 A study from 2003 by Eferl et al27 revealed that the c-JUN protein is required during early stages of tumor development, and in our study, JUN was downregulated at D15.
IGF2 is involved in tissue differentiation. By facilitating the recruitment of transcriptional coactivators, IGF2 positively regulates myogenic transcription factor MYOD1, thus controlling muscle terminal differentiation.28 It has been shown that elevated levels of serum IGF2, which is encoded by the IGF2 gene, have been proven to have an increased risk of developing certain cancers, including breast, lung and colorectal cancers.29 Given that IGF2 was downregulated at D8, this indicates that the proliferation of CRC cells, regarding IGF2 expressivity, decreased. Successive activation of CASP8 and CASP9 play a crucial role in the execution-phase of cell apoptosis.29 It was observed that both genes were not significant in our control data at any time. In our experimental data, the upregulation of CASP8 and CASP9, as well as the downregulation of JUN and IGF2, provide strong evidence that apoptosis increased in KRAS mutated CRC post-reovirus treatment.
MicroRNAs (miRNAs) consisting of small non-coding RNAs are utilized in post-transcriptional gene regulation, many of which are involved in T lymphocyte development, differentiation, and function.30 T cell activation and differentiation are dependent on the expression of multiple miRNAs with many mRNA targets.31 Our study has highlighted several genes within the miR in Lymphocytes gene pathway, which have shown significant alteration post-reovirus treatment.
The CHORDC1 gene regulates the centrosome cycle via ROCKII Kinase to control genomic stability.32 CHORDC1 acts as an onco-suppressor in certain capacities. When under-expressed, CHORDC1 has been shown to induce centrosome amplification and cytogenic abnormalities.33 This amplification is largely caused by increased interactions between ROCKII and NPM, leading to a promotion of ROCK II kinase activity.34 As such, up-regulation of CHORDC1 prevents centrosome amplification and tumorigenesis. Our data indicate that CHORDC1 was upregulated 48 hr, and increased approximately three-fold by D15.
RTN4 plays an important role in the formation and stabilization of endoplasmic reticulum tubules.35 The family of proteins encoded by this gene influences nuclear envelope expansion, nuclear pore complex formation, and proper localization of inner nuclear membrane proteins.36 RTN4B expression in various tissues also regulates angiogenesis, macrophage movement, vascular modeling, sphingolipid homeostasis, inflammation, and other immune responses.37 There are data to indicate that the RTN gene family reduces the anti-apoptotic activity of Bcl-xl and Bcl-2.38 Another isoform of this gene, RTN4-C, is known to be essential in hepatocellular carcinoma regulation, inhibiting cell growth and promoting apoptosis. Our data showed RTN4 to be significantly upregulated at 48 hr indicating its role as an antitumor effector.
FAM96A a component of the cytosolic iron-sulfur protein assembly (CIA) complex has been identified as a novel pro-apoptotic tumor suppressor in gastrointestinal stromal tumors.39 Post-reovirus treatment, our data exhibits significant upregulation of FAM96A at 48 hr.
MIR16-2 is classified within a group of small RNA genes that regulate gene expression.40 The MIR-16 gene acts as a tumor suppressor in cancer cells by inhibiting FEAT translation.41 Decreased MIR-16 expression upregulates expression of MEK1, which in turn promotes cancer cell invasion and proliferation.42 Overexpression of MIR-16 downregulates the ERK/MAPK pathway by inhibiting MEK1 expression. Consequently, this reduces clone formation and inhibits cell proliferation. As our data shows, MIR 16–2 was upregulated at D15. All in all, based on the results from the PBMC samples, lymphocytic maturation and activity increased significantly.
The mitogen-activated protein kinase pathway (MAPK/ERK) and PI3K-Akt pathway play an integral role in CRC, dealing with regulation of gene expression, cellular maturation, and survival.43 Given that reovirus replicates remarkably well within the RAS signaling pathways, our data indicate that after initial reovirus treatment, expression of RAS-specific genes such as RRAS immediately increase in gene expression. This effect occurs because of virus propagation within KRAS mutated cells. However, these genes become downregulated or lose significance as the immune response begins to take effect at D8. The expression of the RRAS gene in our data is a premier example of this phenomenon. At 48 hr, RRAS is upregulated, but loses significance at subsequent timepoints.
PIK3CB is a gene known to be important in signaling pathways that involve receptors on the outer membrane of eukaryotic cells. Ablation of PIK3CB has been shown to impede tumorigenesis with a concomitant diminution of Akt phosphorylation.44 In our study, PIK3CB was shown to be upregulated both at D8 and D15.
NOS3 is a gene that is implicated in vascular smooth muscle relaxation through its production of nitric oxide (NO) via a cGMP-mediated signal transduction pathway.45 Through the activation of platelets, NO mediates VEGF-induced angiogenesis in coronary vessels and promotes blood clotting. Tumor‐derived NOS3 has been shown to promote tumor growth and metastasis by stimulation of tumor cell migration, invasiveness, and angiogenesis. As our data shows, NOS3 was downregulated at D15.
Conclusion
This is a valuable and informative study, with CRC patients receiving reovirus therapy. This study confirms that reovirus (commercially known as pelareorep) has a profound effect on gene expression in patients with KRAS mutated CRC. We have previously shown using the same transcriptome data set that immune related genes8,46 and autophagy-related genes47 are significantly upregulated upon reovirus treatment. Herein, we have further analyzed and reported the genes that are altered in seven important cellular pathways, including the RAS signaling pathway. With deference to genes that displayed significant alteration at multiple timepoints, it was observed that six genes were significant at the 48 hr, 8 were significant at D8, and 10 were significant at D15. Additionally, with respect to genes which were altered in multiple pathways, four genes were significant in the RAS Signaling pathway, four in the MAPK Signaling pathway, three in the EGFR Signaling pathway, six in the PI3K-Akt Signaling pathway, eight in the Angiogenesis pathway, five in the miR-targeted genes in lymphocytes pathway, and five in the Apoptosis pathway. Our transcriptome assay has shown that reovirus treatment causes meaningful alterations in gene expression which indicates that reovirus can be a successful therapy for KRAS mutated CRC. When studied at the transcript level, we are confident, upon comparison with the control data, that administering reovirus in combination with chemotherapy leads to the regression of tumor mediators. Our findings strongly indicate that reovirus treatment leads to immune responses, which include increased lymphocytic maturation and activity, increased levels of apoptosis, and a decrease in angiogenesis. Finally, our data show that while reovirus is propagating, NF-κB, SOS1 and RRAS promote this immune response.
Acknowledgments
We gratefully acknowledge the contribution of the patients and their family members in participating in the clinical trial, which made this study possible. We are thankful to genomic facility of Albert Einstein College of Medicine for performing transcriptome assay. We would like to thank Dr Matt Coffey (Oncolytics Biotech, Calgary, Canada) and Dr Selma Botman, the Yeshiva University Provost and Vice President for Academic Affairs, for graciously providing funding to RM for this research.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval for the version to be published, and agree to be accountable for all aspects of the work. Conception and design: SG, RM. Analysis and interpretation of data: EF, AS, RM. Writing, review, and/or revision of the manuscript: EF, AS, SG, RM. Administrative, technical, or material support: SG, RM. Study supervision: SG, RM.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169(7):1187–1200. doi:10.1016/j.cell.2017.05.045
2. Laffleur B, Basu U. Biology of RNA surveillance in development and disease. Trends Cell Biol. 2019;29(5):428–445. doi:10.1016/j.tcb.2019.01.004
3. Coelho SM, Peters AF, Charrier B, et al. Complex life cycles of multicellular eukaryotes: new approaches based on the use of model organisms. Gene. 2007;406(1–2):152–170. doi:10.1016/j.gene.2007.07.025
4. Nagalakshmi U, Wang Z, Waern K, et al. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320(5881):1344–1349. doi:10.1126/science.1158441
5. Bainbridge MN, Warren RL, Hirst M, et al. Analysis of the prostate cancer cell line LNCaP transcriptome using a sequencing-by-synthesis approach. BMC Genomics. 2006;7(1):246. doi:10.1186/1471-2164-7-246
6. Surget-Groba Y, Montoya-Burgos JI. Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 2010;20(10):1432–1440. doi:10.1101/gr.103846.109
7. Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi:10.1093/bioinformatics/bts635
8. Goel S, Ocean AJ, Parakrama RY, et al. Elucidation of pelareorep pharmacodynamics in a phase I trial in patients with KRAS-mutated colorectal cancer. Mol Cancer Ther. 2020;19(5):1148–1156.
9. Maitra R, Ghalib MH, Goel S. Reovirus: a targeted therapeutic–progress and potential. Mol Cancer Res. 2012;10(12):1514–1525. doi:10.1158/1541-7786.MCR-12-0157
10. Maitra R, Seetharam R, Tesfa L, et al. Oncolytic reovirus preferentially induces apoptosis in KRAS mutant colorectal cancer cells, and synergizes with irinotecan. Oncotarget. 2014;5(9):2807–2819. doi:10.18632/oncotarget.1921
11. Norman KL, Coffey MC, Hirasawa K, et al. Reovirus oncolysis of human breast cancer. Hum Gene Ther. 2002;13(5):641–652. doi:10.1089/10430340252837233
12. Strong JE, Coffey MC, Tang D, et al. The molecular basis of viral oncolysis: usurpation of the Ras signaling pathway by reovirus. EMBO J. 1998;17(12):3351–3362. doi:10.1093/emboj/17.12.3351
13. Lal R, Harris D, Postel-Vinay S, et al. Reovirus: rationale and clinical trial update. Curr Opin Mol Ther. 2009;11(5):532–539.
14. Gollamudi R, Ghalib MH, Desai KK, et al. Intravenous administration of Reolysin, a live replication competent RNA virus is safe in patients with advanced solid tumors. Invest New Drugs. 2010;28(5):641–649. doi:10.1007/s10637-009-9279-8
15. Duncan MR, Stanish SM, Cox DC. Differential sensitivity of normal and transformed human cells to reovirus infection. J Virol. 1978;28(2):444–449. doi:10.1128/jvi.28.2.444-449.1978
16. Duursma AM, Agami R. Ras interference as cancer therapy. Semin Cancer Biol. 2003;13(4):267–273. doi:10.1016/S1044-579X(03)00040-3
17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
18. Hayden MS, West AP, Ghosh S. NF-kappaB and the immune response. Oncogene. 2006;25(51):6758–6780. doi:10.1038/sj.onc.1209943
19. Chardin P, Camonis J, Gale N, et al. Human Sos1: a guanine nucleotide exchange factor for ras that binds to GRB2. Science. 1993;260(5112):1338–1343. doi:10.1126/science.8493579
20. Modzelewska K, Elgort MG, Huang J, et al. An activating mutation in sos-1 identifies Its Dbl domain as a critical inhibitor of the epidermal growth factor receptor pathway during Caenorhabditis elegans vulval development. Mol Cell Biol. 2007;27(10):3695–3707. doi:10.1128/MCB.01630-06
21. Nishida N, Yano H, Nishida T, et al. Angiogenesis in cancer. Vasc Health Risk Manag. 2006;2(3):213–219. doi:10.2147/vhrm.2006.2.3.213
22. Satoh N, Yamada Y, Kinugasa Y, et al. Angiopoietin-1 alters tumor growth by stabilizing blood vessels or by promoting angiogenesis. Cancer Sci. 2008;99(12):2373–2379. doi:10.1111/j.1349-7006.2008.00961.x
23. King SJ, Nowak K, Suryavanshi N, et al. Nesprin-1 and nesprin-2 regulate endothelial cell shape and migration. Cytoskeleton (Hoboken). 2014;71(7):423–434. doi:10.1002/cm.21182
24. Yin M, Yan J, Wei S, et al. CASP8 polymorphisms contribute to cancer susceptibility: evidence from a meta-analysis of 23 publications with 55 individual studies. Carcinogenesis. 2010;31(5):850–857. doi:10.1093/carcin/bgq047
25. Maki Y, Bos TJ, Davis C, et al. Avian sarcoma virus 17 carries the jun oncogene. Proc Natl Acad Sci U S A. 1987;84(9):2848–2852. doi:10.1073/pnas.84.9.2848
26. Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18(1):188–197. doi:10.1093/emboj/18.1.188
27. Eferl R, Ricci R, Kenner L, et al. Liver tumor development. c-Jun antagonizes the proapoptotic activity of p53. Cell. 2003;112(2):181–192. doi:10.1016/S0092-8674(03)00042-4
28. Wilson EM, Rotwein P. Control of MyoD function during initiation of muscle differentiation by an autocrine signaling pathway activated by insulin-like growth factor-II. J Biol Chem. 2006;281(40):29962–29971. doi:10.1074/jbc.M605445200
29. Livingstone C. IGF2 and cancer. Endocr Relat Cancer. 2013;20(6):R321–R339. doi:10.1530/ERC-13-0231
30. de Yebenes VG, Ramiro AR. MicroRNA activity in B lymphocytes. Methods Mol Biol. 2010;667:177–192.
31. Podshivalova K, Salomon DR. MicroRNA regulation of T-lymphocyte immunity: modulation of molecular networks responsible for T-cell activation, differentiation, and development. Crit Rev Immunol. 2013;33(5):435–476. doi:10.1615/CritRevImmunol.2013006858
32. Ferretti R, Sbroggiò M, Di Savino A, et al. Morgana and melusin: two fairies chaperoning signal transduction. Cell Cycle. 2011;10(21):3678–3683. doi:10.4161/cc.10.21.18202
33. Di Savino A, Panuzzo C, Rocca S, et al. Morgana acts as an oncosuppressor in chronic myeloid leukemia. Blood. 2015;125(14):2245–2253. doi:10.1182/blood-2014-05-575001
34. Ferretti R, Palumbo V, Di Savino A, et al. Morgana/chp-1, a ROCK inhibitor involved in centrosome duplication and tumorigenesis. Dev Cell. 2010;18(3):486–495. doi:10.1016/j.devcel.2009.12.020
35. Wang S, Tukachinsky H, Romano FB, et al. Cooperation of the ER-shaping proteins atlastin, lunapark, and reticulons to generate a tubular membrane network. Elife. 2016;5:e18605.
36. Christodoulou A, Santarella-Mellwig R, Santama N, et al. Transmembrane protein TMEM170A is a newly discovered regulator of ER and nuclear envelope morphogenesis in human cells. J Cell Sci. 2016;129(8):1552–1565.
37. Pathak GP, Shah R, Kennedy BE, et al. RTN4 knockdown dysregulates the AKT pathway, destabilizes the cytoskeleton, and enhances paclitaxel-induced cytotoxicity in cancers. Mol Ther. 2018;26(8):2019–2033. doi:10.1016/j.ymthe.2018.05.026
38. Tagami S, Eguchi Y, Kinoshita M, et al. A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene. 2000;19(50):5736–5746. doi:10.1038/sj.onc.1203948
39. Schwamb B, Pick R, Fernández SBM, et al. FAM96A is a novel pro-apoptotic tumor suppressor in gastrointestinal stromal tumors. Int J Cancer. 2015;137(6):1318–1329. doi:10.1002/ijc.29498
40. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
41. Liang H, Fu Z, Jiang X, et al. miR-16 promotes the apoptosis of human cancer cells by targeting FEAT. BMC Cancer. 2015;15(1):448. doi:10.1186/s12885-015-1458-8
42. Chen T, Xiao Q, Wang X, et al. miR-16 regulates proliferation and invasion of lung cancer cells via the ERK/MAPK signaling pathway by targeted inhibition of MAPK kinase 1 (MEK1). J Int Med Res. 2019;47(10):5194–5204. doi:10.1177/0300060519856505
43. Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. doi:10.1517/14728222.2011.645805
44. Jia S, Liu Z, Zhang S, et al. Essential roles of PI(3) K-p110betain cell growth, metabolism and tumorigenesis. Nature. 2008;454(7205):776–779. doi:10.1038/nature07091
45. Francis SH, Busch JL, Corbin JD, et al. cGMP-dependent protein kinases and cGMP phosphodiesterases in nitric oxide and cGMP action. Pharmacol Rev. 2010;62(3):525–563. doi:10.1124/pr.110.002907
46. Parakrama R, Fogel E, Chandy C, et al. Immune characterization of metastatic colorectal cancer patients post reovirus administration. BMC Cancer. 2020;20(1):569. doi:10.1186/s12885-020-07038-2
47. Jiffry J, Thavornwatanayong T, Rao D, et al. Oncolytic Reovirus (pelareorep) induces autophagy in KRAS-mutated colorectal cancer. Clin Cancer Res. 2021;27(3):865–876. doi:10.1158/1078-0432.CCR-20-2385
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



