Back to Journals » Journal of Pain Research » Volume 13
Transcriptome Analysis of Dorsal Root Ganglion in Rats with Knee Joint Inflammation
Authors Bai Q , Cao J, Dong T, Tao F
Received 24 August 2020
Accepted for publication 7 October 2020
Published 28 October 2020 Volume 2020:13 Pages 2709—2720
DOI https://doi.org/10.2147/JPR.S278474
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Robert B. Raffa
Qian Bai,1 Jing Cao,2 Tieli Dong,1 Feng Tao3
1Department of Anesthesiology, The Second Affiliated Hospital of Zhengzhou University, Henan, People’s Republic of China; 2Department of Anatomy, School of Basic Medical Sciences, Zhengzhou University, Henan, People’s Republic of China; 3Department of Biomedical Sciences, Texas A&M University College of Dentistry, Dallas, Texas, USA
Correspondence: Tieli Dong
The Second Affiliated Hospital of Zhengzhou University, Henan, People’s Republic of China
Email [email protected]
Feng Tao
Texas A&M University College of Dentistry, Dallas, Texas, USA
Email [email protected]
Background: Rheumatoid arthritis (RA) leads to pain through alteration of gene expression. Although gene expression alteration in knee cartilage or peripheral blood from RA patients has been identified using microarray, it remains unclear whether long non-coding RNA (lncRNA)-mediated gene regulation occurs in primary sensory neurons of dorsal root ganglia (DRG) during RA-like joint inflammation. In the present study, we aimed to analyze lncRNA and related mRNA profiles in the DRG in a knee joint inflammation rat model.
Methods: Complete Freund’s adjuvant (CFA) was injected in the rat knee joint for preparing the joint inflammation model. A lncRNA-mRNA microarray of rat DRG was employed for transcriptome analysis. Functional roles of differentially expressed lncRNAs and their related mRNAs in the injured DRG were delineated by bioinformatic analysis.
Results: We observed that expression levels of 9000 lncRNAs were altered on day 7 post-CFA, of which 45.17% were up-regulated and 54.83% were down-regulated. Specifically, 69 lncRNAs (42 up and 27 down) were significantly regulated. We also observed that expression levels of 13,744 mRNAs were altered on day 7 post-CFA, of which 49.67% were up-regulated and 50.33% were down-regulated. Specifically, 102 mRNAs (51 up and 51 down) were significantly regulated. Using quantitative real-time PCR, we verified the changes in differentially expressed lncRNAs in the injured DRG.
Conclusion: These results suggest that microarray-based RNA sequencing can be used to identify altered lncRNAs and relevant mRNAs in the DRG of rats with knee joint inflammation.
Keywords: joint inflammation, dorsal root ganglion, transcriptome analysis
Introduction
Patients with rheumatoid arthritis (RA) suffer from chronic widespread pain and chronic regional pain. These symptoms significantly decrease their quality of life.1 However, current therapies for chronic knee pain are either not effective or with serious side effects.2 Thus, it is urgent to understand the underlying mechanisms for developing a new therapy for such pain.
Long non-coding RNAs (lncRNAs) are transcribed RNA molecules with no or very low protein-coding potential.3 Numerous lncRNAs have been identified to exert vital parts in regulating the expression of specific genes and are involved in different biological functions.4,5 The abnormal expression of lncRNAs will result in different neurological disorders. Previous studies have shown that changes in the expression of lncRNAs in dorsal root ganglia (DRG) neurons have been implicated in the pathogenesis of neuropathic pain.6–11 Gene expression alteration in knee cartilage, synovium or peripheral blood from RA patients have been identified using microarray,12–15 but it remains unclear whether lncRNAs-mediated gene regulation occurs in primary sensory neurons of DRG during RA-like joint inflammation.
In this study, we analyzed lncRNAs and related mRNAs in the DRG after knee joint inflammation induction. A unilateral intra-articular injection of complete Freund’s adjuvant (CFA) in rats was performed to prepare a rodent model of joint inflammation. Using microarray RNA sequencing, we identified 69 lncRNAs and 102 related mRNAs that displayed significant expression alteration in the DRG after knee joint inflammation. These lncRNAs and mRNAs could be potential targets for developing novel therapies for inflammatory joint pain. As indicated by the technical validation in this study, high-quality data were obtained, and the expression levels of genes took place within the desired regions of genes. Our findings provide precious data for the scientific community to more intensively investigate the alterations of DRG at molecular level in the CFA-induced inflammatory joint pain model.
Materials and Methods
Animals
Male inbred Wistar rats (200 g) were provided by Animal Laboratories Center of Zhengzhou University in China and were housed in the central facility at Zhengzhou University School of Medicine under a standard 12 h light/12 h dark cycle, with water and food available adlibitum. The study protocol gained approval from the Institutional Ethics Review Committee at Zhengzhou University. Each experiment was performed according to the guidelines for the care and use of laboratory animals released by the National Institutes of Health (NIH). The animal number utilized together with their sufferings was minimized as far as possible. The reviewers were blinded to animal treatment.
CFA-Induced Inflammatory Joint Pain Rat Model
Three percent isoflurane was injected into rats for anesthesia, then 150 μL CFA (1 mg/mL Mycobacterium tuberculosis, Sigma, USA) was also injected into their right knees via the patella tendon. We prepared the animal model as described previously.16 As a control, 150 μL of saline (0.9% sodium chloride) was injected into the right knee joint. Rats were allowed to recover from anesthesia before being returned to their cages.
Determination of Rat Knee Diameter
For animals under anesthesia, their knee joint diameters just below the level of the patella were determined using a digital vernier caliper (ID:110–601, Langir Electric CO., Ltd.) as described previously.16 On the 7th day following intra-articular injection with CFA, the edema in knee was quantified.
Pain Behavioral Test
Mechanical behavioral testing (6 rats per group from a different set of rats) was performed according to our prior articles.17–19 The up–down testing paradigm was employed to measure the 50% paw withdrawal threshold (PWT) responding to the mechanical stimulus by von Frey filaments (North Coast Medical Inc., Gilroy, CA, USA), according to the previous description.18,19
RNA Extraction
Right L4 and L5 DRGs from CFA group and saline control group were harvested on day7 after CFA or saline injection (3 rats per group from a different set of rats). Next, the mini RNA easy kit (QIAGEN, Valencia, CA, USA) was used to extract total RNA, and then genomic DNA was digested in the column in accordance with specific protocols. Thereafter, NanoDrop ND-1000 (Thermo Scientific, Wilmington, DE, USA) was used to measure RNA quality and quantity, whereas the standardized denatured AGE was applied in assessing the RNA integrity.
Microarray Analysis
A lncRNA array of rats was prepared to profile the lncRNAs as well as the protein-coding genes. Meanwhile, gene-coding probes were printed for once, whereas the lncRNAs-coding ones were printed for thrice, and negative ones together with housekeeping genes-coding ones were printed several times to ensure the quality of hybridization. The slightly modified Agilent One-Color Microarray-Based Gene Expression Analysis protocol (Agilent Technology) was used to label samples and hybridize the array. In brief, rRNA was removed from total RNA using the mRNA-ONLY™ Eukaryotic mRNA Isolation Kit (Epicentre), followed by mRNA purification. Subsequently, all samples were subjected to amplification and transcription to the fluorescence cRNA down the full-length transcripts in the absence of 3ʹ bias according to the random priming approach (Arraystar Flash RNA Labeling Kit, Arraystar). Then, the RNeasy Mini Kit (Qiagen) was used to purify those labeled cRNAs. The NanoDrop ND-1000 was used to measure labeled cRNAs content along with the activity. Afterward, 10 × blocking reagent (5 μL) together with 25 × fragmentation buffer (1 μL) were added to fragment the labeled cRNAs (1 μg each), followed by 30 min heating under 60 °C. At last, those labeled cRNAs were diluted by 2 × GE hybridization buffer (25 μL). Later, the hybridization solution (50 μL) was added to the gasket slide for the assembly of lncRNA expression microarray slides. Then, each slide was incubated within the Agilent Hybridization Oven under 65 °C for 17 h. Afterward, an Agilent DNA Microarray Scanner (Agilent, part # G2505C) was utilized to wash, fix, and scan those hybridized arrays.
Data Processing
Altogether six samples were used for sequencing, multiplexing, analysis for differentially expressed (DE) genes, as well as analysis of expression of transcripts and lncRNAs. In brief, the array images were analyzed by Agilent Feature Extraction (version 11.0.1.1). The GeneSpring GX v12.1 (Agilent Technologies) was employed to normalize quantiles and process data. When the raw data were normalized for their quantiles, mRNAs and lncRNAs with flags in ≥3 among the six samples in Marginal or Present (“All Targets Value”) were selected to analyze data. Then, the significant DE lncRNAs and DE mRNAs between CFA injection and normal control groups were discovered according to the same threshold of P-value as that in screening DE lncRNAs and DE mRNAs. The data were submitted to NCBI Gene Expression Omnibus (GEO: GSE146881). This GEO project includes raw data for all samples.
Enrichment and Pathway Analysis
GO analysis facilitates to interpret the properties of genes and their products within an organism (http://www.geneontology.org). We used top GO for GO analysis of differential mRNAs to infer their biological functions: 1) Biological Processes (BPs); 2) Molecular Functions (MFs); 3) Cellular Components (CCs). The KEGG pathway analysis was performed for identifying those biological pathways related to DE mRNAs.20
Quantitative Real-Time PCR
The ThermoScript reverse transcriptase (Invitrogen) was utilized to reversely transcribe total RNA in accordance with specific protocols. Table 1 lists the primers used to amplify cDNA through qRT-PCR. All samples were examined for three times by the BIO-RAD CFX96 RT-PCR system. The PCR conditions were shown below: 3 min of initial denaturation under 95°C; 10 s under 95°C, 30 s under 60°C, 30 s under 72°C for 40 cycles. The ipsilateral to contralateral lncRNA expression ratio was determined by the standardized 2−∆∆Ct method. GAPDH was used as the internal reference.
 |
Table 1 Primers for RT-qPCR |
Establishment of the Co-Expression Network
Pearson correlation calculation (PCC) was utilized to construct the coding-noncoding gene co-expression (CNC) network to normalize the DE lncRNAs signal intensity relative to that of DE mRNAs. The results of PCC > 0.9 were screened as interrelationship pairs. We used the Cytoscape (v2.8.1) to draw a CNC network.
Statistical Analysis
All values were presented in the manner of means ± SEM. SPSS 17.0 was employed for all statistical analyses. Differences in specific lncRNAs or mRNAs levels between CFA injection and saline control groups discovered from microarray as well as PCR were analyzed by Student’s t-test. A difference of two-sided p < 0.05 indicated statistical significance.
Results
CFA-Induced Inflammatory Joint Pain
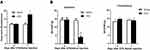
In the CFA-induced inflammatory joint pain rat model, we observed that unilateral injection of CFA into the knee joint not only significantly increased knee joint diameter (Figure 1A) but also markedly decreased PWT (Figure 1B) at the ipsilateral side on day 7 post-injection, which indicates intra-articular injection of CFA induces inflammatory joint pain. Our observation is consistent with previous studies.16,21
Expression Profiles of lncRNAs and mRNAs in the DRGs After CFA Injection
On the 7th day following CFA treatment, a graphical overview of the expression signatures of lncRNAs and mRNAs in the L4 and L5 DRGs were obtained by using scatter plot and hierarchical cluster analyses of the microarray results. As observed from the scatter plot, numerous mRNAs and lncRNAs showed differential expression in CFA treatment group compared with saline control group (Figure 2A and B). Besides, it was illustrated from the hierarchical clustering analysis on mRNAs and lncRNAs that, those respective three samples under saline or CFA treatment were clustered, with identical signal intensity in CFA or saline control group (Figure 2C and D). The heatmap showed that DE lncRNAs or mRNAs that were either up-regulated or down-regulated twofold (Figure 2E and F), indicating the high level of concordance in both saline-treated and CFA-treated samples.
DE lncRNAs and mRNAs Following RA Induction
We further analyzed DE lncRNAs and mRNAs using the significance analysis of microarray. Totally, 69 DE lncRNAs, including 42 up-regulated and 27 down-regulated, were significantly altered in the CFA-treated group compared with the saline control group. lncRNAs with the highest up-regulation level included MRAK139263, uc.247+, MRAK163594, AY539883, MRAK081329, and MRAK081329, of which MRAK139263 showed the largest up-regulation (log2 fold change = 4.3602434). lncRNAs with the highest down-regulation level included XR_005599, uc.288+, AY321352, uc.83+ and uc.344-, of which XR_005599 showed the largest down-regulation (log2 fold change = −3.6773075). The detailed information about the top 20 up-regulated and 20 down-regulated lncRNAs is listed in Table 2. For the DE mRNAs, 51 up-regulated mRNAs and 51 down-regulated mRNAs were identified in the CFA-induced RA group compared with the saline control group. Those DE mRNAs contained multiple known genes related to the processing of pain, such as 1) Scn2a1 (encodes NaV1.2, a voltage-dependent sodium channel), 2) C1qtnf3 (encodes C1q/TNF-related protein 3, and 3) Mpo (encodes myeloperoxidase). In addition, several genes with unknown roles in pain also showed significant alteration following RA induction, including Ampd1 (encodes adenosine monophosphate deaminase 1) and S100a8 (encodes S100 calcium-binding protein A8). The detailed information about the top 20 up-regulated and 20 down-regulated mRNAs are listed in Table 3.
 |
Table 2 The Detail Information of the Top 20 Up-Regulated and 20 Down-Regulated lncRNAs |
 |
Table 3 The Detail Information of the Top 20 uUp-Regulated and 20 Down-Regulated mRNAs |
Class Distribution Analysis of DE lncRNAs
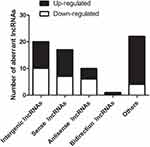
LncRNAs are shown previously to modulate overlapping or adjacent mRNAs expression within the genome.22–24 In this study, all DE lncRNAs were classified into five categories according to their genomic proximity to protein-coding genes: 1) intergenic lncRNAs without a coding transcript in the 30 kb range of lncRNAs; 2) sense lncRNAs whose exons were overlapping with the exons of coding transcripts or introns in the coding transcripts in an identical genomic strand; 3) antisense lncRNAs transcribed based on an antisense strand, which overlapped with the coding transcripts or did not share the overlapping exons; 4) bidirectional lncRNAs directed head-to-head to the coding transcripts in the range of 1000 bp; 5) other lncRNAs. In the 69 DE lncRNAs modulated by CFA-induced joint inflammation, we identified that 20 Intergenic lncRNAs, 17 Sense lncRNAs, 10 Antisense lncRNAs, 1 Bidirectional lncRNAs, and 22 other types of lncRNAs (Figure 3).
Quantitative Real-Time PCR Validation of lncRNA Expression
To validate the results of microarray, the up-regulated lncRNAs (MRAK163594 and uc.247+) and the down-regulated lncRNAs (uc.83-, XR_005599, NONRATT021972, and AY321352) were analyzed by quantitative real-time PCR. The DRG tissues were harvested for assessing lncRNA expression on day 7 after intra-articular injection of CFA or saline. We observed that MRAK163594 and uc.247+ were significantly increased in CFA-treated rats in comparison with normal controls (Figure 4A and B) and that uc.83-, XR_005599, NONRATT021972, and AY321352 were remarkably declined in CFA-treated rats in comparison with normal controls (Figure 4C−F). Those markedly changed lncRNAs were used to construct the CNC network (Figure 4G).
Functional Prediction of DE mRNAs in Inflammatory Joint Pain
To explore the molecular mechanism of inflammatory joint pain, we conducted GO and KEGG enrichment analysis of regulated genes in the CFA-induced inflammatory joint pain rat model. Using GO, we showed that 1) the most significantly enriched molecular functions of the up-regulated genes included protein complex binding, exonuclease activity, and antioxidant activity (Figure 5A), 2) the most significantly enriched biological processes of the up-regulated genes included defense response, response to external stimulus, and response to stress (Figure 5B), 3) the most markedly enriched cellular components of the up-regulated genes included extracellular space, extracellular region part, and extracellular region (Figure 5C), 4) the most significantly enriched molecular functions of the down-regulated genes included alcohol binding, transporter activity, and calcium channel binding (Figure 5D), 5) the most significantly enriched biological processes of the down-regulated genes included muscle organ development, muscle structure development, and skeletal muscle tissue development (Figure 5E), and 6) the most markedly enriched cellular components of the down-regulated genes included extracellular region, extracellular space, extracellular region part, and cell surface (Figure 5F).
Using KEGG, we showed that 1) the up-regulated genes were involved in inflammatory mediators-produced regulation of transient receptor potential channels, nicotine addiction, intestinal immune network for IgA production, and vitamin B6 metabolism (Figure 6A), and 2) the down-regulated genes were involved in hypertrophic cardiomyopathy, peroxisome proliferator-activated receptor (PPARs) signaling pathway, nitrogen metabolism, and drug metabolism-cytochrome P450 (Figure 6B).
Discussion
Inflammatory joint pain is associated with gene expression changes within the DRG. Identifying altered genes is critical for defining the mechanisms underlying inflammatory joint pain pathogenesis. In the present study, we conducted the Rat lncRNA and mRNA array to reveal transcriptome alterations after induction of inflammatory joint pain with CFA.
We used quantitative real-time PCR to verify the expression of six randomly selected DE lncRNAs in the DRG on day 7 after CFA injection, including up-regulated lncRNAs MRAK163594 and uc.247+ and down-regulated lncRNAs uc.83-, XR_005599, NONRATT021972, and AY321352. We constructed a CNC network for the most significantly changed lncRNAs. In the CNC network, the up-regulated MRAK163594 and the down-regulated uc.83- were co-expressed with known gene Kcnj16,25 and the up-regulated MRAK163594 and uc.247+ were co-expressed with pain-related gene Scn2a,26 which contributes to many pain conditions, such as inflammatory joint pain.21,27 In addition, a previous study has shown that lncRNA uc.217 regulates neurite outgrowth in DRG neurons following peripheral nerve injury.28 By co-expressing with Scn9a, the lncRNA uc.217 is likely to play an important role in CFA-induced inflammatory joint pain.21
Furthermore, the identified DE mRNAs in our study contain several known genes involved in pain processing. 1) Scn2a (encodes NaV1.2, a voltage-dependent sodium channel): this ion channel has been demonstrated to regulate pain conduction;26 2) C1qtnf3(encodes C1q/TNF-related protein 3): this protein plays an important role in the development of RA);29 3) Mpo (encodes myeloperoxidase): the enzyme activity is an index of neutrophilic granulocyte infiltration, which has been recognized as a marker for pain.30,31
On the other hand, the identified DE mRNAs also contain several genes with unknown roles in pain. 1) Ampd1 (encodes adenosine monophosphate deaminase 1): Patients with Ampd1 deficiency experience muscle pain (myalgia) or weakness after exercise;32 2) S100a8 (encodes S100 calcium-binding protein A8): the S100a8 protein is positively associated with increased knee symptoms, cartilage defects in patients with knee osteoarthritis.33 In addition, KEGG pathway analysis for the DE mRNAs indicates some potential targets, such as inflammatory mediator-regulated transient receptor potential channels, which have been demonstrated as important molecules involved in joint inflammation in the DRG.34
Conclusion
Our results suggest that aberrantly expressed lncRNAs contribute to the development of inflammatory joint pain.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding authors on reasonable request.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed on the journal to which the article will be submitted; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Henan Medical Science and Technology Research Youth Project Co-Sponsored by the Province and Ministry in China (Grant # SB201902020) and Top Talent Fund of the Second Affiliated Hospital of Zhengzhou University (Grant# 2020BJRCA03).
Disclosure
The authors declare that there are no conflicts of interest.
References
1. Andersson ML, Svensson B, Bergman S. Chronic widespread pain in patients with rheumatoid arthritis and the relation between pain and disease activity measures over the first 5 years. J Rheumatology. 2013;40(12):1977–1985. doi:10.3899/jrheum.130493
2. van Laar M, Pergolizzi JV
3. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
4. Jandura A, Krause HM. The new RNA world: growing evidence for long noncoding RNA functionality. Trends Genetics. 2017;33(10):665–676. doi:10.1016/j.tig.2017.08.002
5. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Molecular Cell. 2011;43(6):904–914. doi:10.1016/j.molcel.2011.08.018
6. Baskozos G, Dawes JM, Austin JS, et al. Comprehensive analysis of Long non-coding RNA expression in dorsal root ganglion reveals cell type specificity and dysregulation following nerve injury. Pain. 2018.
7. Jiang BC, Sun WX, He LN, Cao DL, Zhang ZJ, Gao YJ. Identification of lncRNA expression profile in the spinal cord of mice following spinal nerve ligation-induced neuropathic pain. Mol Pain. 2015;11:43. doi:10.1186/s12990-015-0047-9
8. Liu Z, Liang Y, Wang H, et al. LncRNA expression in the spinal cord modulated by minocycline in a mouse model of spared nerve injury. J Pain Res. 2017;10:2503–2514. doi:10.2147/JPR.S147055
9. Mao P, Li CR, Zhang SZ, Zhang Y, Liu BT, Fan BF. Transcriptomic differential lncRNA expression is involved in neuropathic pain in rat dorsal root ganglion after spared sciatic nerve injury. Brazilian j Med Biol Res. 2018;51(10):e7113. doi:10.1590/1414-431x20187113
10. Zhou J, Fan Y, Chen H. Analyses of long non-coding RNA and mRNA profiles in the spinal cord of rats using RNA sequencing during the progression of neuropathic pain in an SNI model. RNA Biology. 2017;14(12):1810–1826. doi:10.1080/15476286.2017.1371400
11. Zhou J, Xiong Q, Chen H, Yang C, Fan Y. Identification of the spinal expression profile of non-coding RNAs involved in neuropathic pain following spared nerve injury by sequence analysis. Front Mol Neurosci. 2017;10:91. doi:10.3389/fnmol.2017.00091
12. Luo Q, Xu C, Li X, et al. Comprehensive analysis of long non-coding RNA and mRNA expression profiles in rheumatoid arthritis. Exp Ther Med. 2017;14(6):5965–5973.
13. Fu M, Huang G, Zhang Z, et al. Expression profile of long noncoding RNAs in cartilage from knee osteoarthritis patients. Osteoarthritis Cartilage OARS, Osteoarthritis Research Society. 2015;23(3):423–432. doi:10.1016/j.joca.2014.12.001
14. Bratus-Neuenschwander A, Castro-Giner F, Frank-Bertoncelj M, et al. Pain-associated transcriptome changes in synovium of knee osteoarthritis patients. Genes. 2018;9:7. doi:10.3390/genes9070338
15. Zhang Y, Xu YZ, Sun N, et al. Long noncoding RNA expression profile in fibroblast-like synoviocytes from patients with rheumatoid arthritis. Arthritis Research Therapy. 2016;18(1):227. doi:10.1186/s13075-016-1129-4
16. Martindale JC, Wilson AW, Reeve AJ, Chessell IP, Headley PM. Chronic secondary hypersensitivity of dorsal horn neurones following inflammation of the knee joint. Pain. 2007;133(13):79–86. doi:10.1016/j.pain.2007.03.006
17. Su S, Shao J, Zhao Q, et al. MiR-30b attenuates neuropathic pain by regulating voltage-gated sodium channel nav1.3 in Rats. Frontiers Molecular Neuroscience. 2017;10:126. doi:10.3389/fnmol.2017.00126
18. Cai W, Zhao Q, Shao J, et al. MicroRNA-182 alleviates neuropathic pain by regulating nav1.7 following spared nerve injury in rats. Sci Rep. 2018;8(1):16750. doi:10.1038/s41598-018-34755-3
19. Bai Q, Shao J, Cao J, et al. Protein kinase C-alpha upregulates sodium channel Nav1.9 in nociceptive dorsal root ganglion neurons in an inflammatory arthritis pain model of rat. J Cellular Biochemistry. 2020;121:
20. Kanehisa M, Araki M, Goto S, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Research. 2008;36(Database issue):D480–4. doi:10.1093/nar/gkm882
21. Strickland IT, Martindale JC, Woodhams PL, Reeve AJ, Chessell IP, McQueen DS. Changes in the expression of NaV1.7, NaV1.8 and NaV1.9 in a distinct population of dorsal root ganglia innervating the rat knee joint in a model of chronic inflammatory joint pain. Eur J Pain. 2008;12(5):564–572. doi:10.1016/j.ejpain.2007.09.001
22. Liu C, Li C, Deng Z, Du E, Xu C. Long Non-coding RNA BC168687 is Involved in TRPV1-mediated diabetic neuropathic pain in rats. Neuroscience. 2018;374:214–222. doi:10.1016/j.neuroscience.2018.01.049
23. Wang S, Xu H, Zou L, et al. LncRNA uc.48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signalling. 2016;12(1):139–148. doi:10.1007/s11302-015-9488-x
24. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–U58. doi:10.1038/nature09819
25. D’Adamo MC, Shang L, Imbrici P, Brown SD, Pessia M, Tucker SJ. Genetic inactivation of Kcnj16 identifies Kir5.1 as an important determinant of neuronal PCO2/pH sensitivity. J Biological Chemistry. 2011;286(1):192–198. doi:10.1074/jbc.M110.189290
26. Liao Y, Anttonen AK, Liukkonen E, et al. SCN2A mutation associated with neonatal epilepsy, late-onset episodic ataxia, myoclonus, and pain. Neurology. 2010;75(16):1454–1458. doi:10.1212/WNL.0b013e3181f8812e
27. Schuelert N, McDougall JJ. Involvement of Nav 1.8 sodium ion channels in the transduction of mechanical pain in a rodent model of osteoarthritis. Arthritis Res Ther. 2012;14(1):R5. doi:10.1186/ar3553
28. Yao C, Wang J, Zhang H, et al. Long non-coding RNA uc.217 regulates neurite outgrowth in dorsal root ganglion neurons following peripheral nerve injury. European J Neuroscience. 2015;42(1):1718–1725. doi:10.1111/ejn.12966
29. Murayama MA, Kakuta S, Maruhashi T, et al. CTRP3 plays an important role in the development of collagen-induced arthritis in mice. Biochem Biophys Res Commun. 2014;443(1):42–48. doi:10.1016/j.bbrc.2013.11.040
30. Bhalla S, Singh N, Jaggi AS. Dose-related neuropathic and anti-neuropathic effects of simvastatin in vincristine-induced neuropathic pain in rats. Food Chemical Toxicology. 2015;80:32–40. doi:10.1016/j.fct.2015.02.016
31. Britti D, Crupi R, Impellizzeri D, et al. A novel composite formulation of palmitoylethanolamide and quercetin decreases inflammation and relieves pain in inflammatory and osteoarthritic pain models. BMC Veterinary Research. 2017;13(1):229. doi:10.1186/s12917-017-1151-z
32. Lim L, Palayer M, Bruneau A, et al. Myoadenylate deaminase deficiency: a frequent cause of muscle pain A case detected by exercise testing. Ann Biol Clin (Paris). 2017;75(4):445–449.
33. Ruan G, Xu J, Wang K, et al. Associations between serum S100A8/S100A9 and knee symptoms, joint structures and cartilage enzymes in patients with knee osteoarthritis. Osteoarthritis Cartilage OARS, Osteoarthritis Research Society. 2019;27(1):99–105. doi:10.1016/j.joca.2018.08.020
34. Keeble J, Russell F, Curtis B, Starr A, Pinter E, Brain SD. Involvement of transient receptor potential vanilloid 1 in the vascular and hyperalgesic components of joint inflammation. Arthritis Rheum. 2005;52(10):3248–3256. doi:10.1002/art.21297
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.