Back to Journals » OncoTargets and Therapy » Volume 13
Traditional Chinese Medicine Xiaoai Jiedu Recipe Suppresses the Development of Hepatocellular Carcinoma via Regulating the microRNA-29a/Signal Transducer and Activator of Transcription 3 Axis
Authors Shi Y, Kong W, Lu Y, Zheng Y
Received 8 February 2020
Accepted for publication 15 June 2020
Published 29 July 2020 Volume 2020:13 Pages 7329—7342
DOI https://doi.org/10.2147/OTT.S248797
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Yanfen Shi,1 Wuming Kong,2 Yanxu Lu,1 Yu Zheng3
1Jiaozuo People’s Hospital, Jiaozuo City, Henan Province 454000, People’s Republic of China; 2Department of Gastroenterology, Shanghai University of Medicine & Health Sciences Affiliated Sixth People’s East-Hospital, Shanghai City 201306, People’s Republic of China; 3Kunming Municipal Hospital of Traditional Chinese Medicine, Kunming City, Yunnan, Province 650011, People’s Republic of China
Correspondence: Yu Zheng Department of Spleen and Stomach Disease, No. 25, Dongfeng East Road, Panlong District, Kunming City, Yunnan Province 650011, People’s Republic of China
Tel +86-0871-63804608
Email [email protected]
Objective: Hepatocellular carcinoma (HCC) is one of the most frequent and lethal tumors affecting human health worldwide. The aim of this study was to investigate the anti-cancer effects of Xiaoai Jiedu Recipe (XJR) on HCC development and its underlying mechanisms.
Methods: The expression of microRNA-29a (miR-29a) and signal transducer and activator of transcription 3 (STAT3) in HCC tissues and cells was determined by quantitative real-time polymerase chain reaction. The proliferation, migration, and invasion of HCC cells were measured by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide, wound-healing, and transwell assays, respectively. The regulatory relationship between miR-29a and STAT3 in HCC was predicted by TargetScan and analyzed by luciferase reporter and RNA pull-down assays. The protein expression of matrix metalloproteinase (MMP)-2/9 and STAT3 was detected by Western blotting. A xenograft tumor mouse model was established, and tumor weight and volume were measured.
Results: The expression of miR-29a was significantly decreased in HCC tissues and cells compared with that in normal tissues and cells. The up-regulation of miR-29a was related with lymph node metastasis and tumor node metastasis stage. XJR treatment significantly increased the expression of miR-29a, decreased cell viability, migration, and invasion, and reduced the protein expression of MMP-2/9 in HCC cells in a concentration-dependent manner. The anti-tumor effect of XJR on HCC cells was reversed by treatment with miR-29a inhibitor. STAT3 was predicted as a target of miR-29a, and its expression was negatively regulated by miR-29a. Moreover, STAT3 knockdown suppressed the malignant behavior of HCC cells, and its anti-tumor function was reversed by treatment with miR-29a inhibitor. Furthermore, XJR treatment inhibited tumor growth in mice through elevating miR-29a expression and inhibiting STAT3 expression.
Conclusion: XJR suppressed the development of HCC via regulating miR-29a and STAT3.
Keywords: Xiaoai Jiedu Recipe, hepatocellular carcinoma, microRNA-29a, STAT3, cell behaviors
Introduction
Hepatocellular carcinoma (HCC), one of the most common and fatal tumors, is the leading cause of malignancy-related deaths worldwide, especially in China.1,2 Hepatitis B/C virus infection, cirrhosis, smoking, diabetes, and alcohol abuse are the major reasons for the increasing occurrence rates of HCC.3 At present, surgical resection is the best treatment strategy for HCC.4 However, the outcomes of patients with HCC remain unsatisfactory due to late diagnosis, distant metastasis, and high recurrence rate.5
Traditional Chinese medicine (TCM), a hot spot for tumor treatment, has several advantages, and has been widely used for the treatment of diverse cancer types in China.6,7 For example, Jiedu Sangen Decoction, which has been used in the treatment of colorectal cancer (CRC) for over 50 years, has the ability to suppress the migration and invasion of CRC cells.8,9 In addition, TCM has been long applied for the treatment of HCC. Besides, transcatheter arterial chemoembolization combined with JDF was shown to improve the prognosis of patients with HCC.10 Jie-du Recipe has also been shown to prolong the survival of patients with HCC and prevent recurrence.11 Xiaoai Jiedu Recipe (XJR) is an effective anti-cancer drug that has been long used in clinical practice under the guidance of Professor Zhongying Zhou of Nanjing University of Traditional Chinese Medicine. Zhou et al found that treatment with XJR increases short-term clinical efficacy, reduces the toxicity of chemotherapeutic drugs, and improves the quality of life of patients with advanced cancer.12 In addition, Shi et al showed that XJR treatment decreased tumor volume and weight in colon cancer-bearing mice.13 Furthermore, Chen et al demonstrated that XJR treatment significantly inhibited the growth of H22-transplanted tumors and promoted cell apoptosis in mice.14 However, the specific therapeutic effects of XJR on HCC cells in vitro and the underlying mechanisms are not yet fully understood.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that play important roles in the pathogenesis of HCC.15 miR-29a is a potential diagnostic and prognostic biomarker of HCC.16 Mahati et al demonstrated that miR-29a is down-regulated in HCC tissues and cells, and overexpression of miR-29a inhibits tumor growth and migration in HCC.17 Additionally, Zhang et al showed that low expression of miR-29a is associated with tumor size and vascular invasion, and overexpression of miR-29a suppresses HCC cell proliferation, colony formation ability, and cell cycle progression.18 Notably, an miRNA expression profile of H22-bearing mice showed that XJR treatment significantly up-regulated the expression of miR-1298-5p, −874-3p, −721,-298-5p, −551b-5p, −346-5p, and −105, and down-regulated that of miR-24-3p, −3963, −127-3p, −434-5p, −1187, −468-3p, −221-5p, and −6695-5p.19 Our pre-experiments proved that miR-29a expression was up-regulated by XJR treatment. However, the underlying mechanism by which XJR regulates miR-29a expression in HCC remains unclear.
Signal transducer and activator of transcription 3 (STAT3) is a key signaling molecule involved in malignant transformation.20 STAT3 is usually activated in HCC, and silencing of STAT3 has been considered as a therapeutic strategy for HCC.21 Zuo et al showed that treatment with LLL12, a small inhibitor targeting STAT3, inhibited the proliferation and induced the apoptosis of HCC cells in vitro, as well as inhibited the growth of SNU398 xenografts in mice.22 Furthermore, Jung et al showed that C188-9, a novel small-molecule STAT3 inhibitor, exhibits anti-tumor roles in HCC in vitro and in vivo.23 In addition, STAT3 is a known target of miR-29a in retinoblastoma and nasopharyngeal carcinoma.24,25 However, the regulatory relationship between miR-29a and STAT3 in HCC remains unclear.
In this study, the therapeutic effects of XJR on HCC cells and tissues and the underlying mechanism involving miR-29a/STAT3 were analyzed. Our findings provide insights into the anti-tumor role of XJR and may help in the development of a potential therapeutic strategy for HCC.
Materials and Methods
Patients
A total of 40 patients with HCC and 40 normal volunteers were recruited at Kunming Municipal Hospital of Traditional Chinese Medicine. The clinical characteristics of all recruited patients are listed in Table 1. This study was approved by the Ethics Committee of Kunming Municipal Hospital of Traditional Chinese Medicine and conducted in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients.
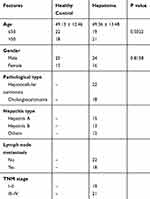 |
Table 1 The Clinical Characteristics of HCC Patients and Healthy Controls |
XJR Preparation
XJR, containing Hedyotis diffusa (20 g), Shanshi mushroom (10 g), Ophiopogon japonicus (12 g), Pseudostellaria heterophylla (15 g), August sturgeon (12 g), stiff silkworm (10 g), and pupae (3 g) was purchased from Bozhou Traditional Chinese and Western Medicine Co., Ltd. (Anhui, China). XJR was identified by a professor in the School of Pharmacy of Nanjing University of Traditional Chinese Medicine according to the 2015 edition of the Pharmacopoeia of the People’s Republic of China. XJR was concentrated to 2.0 g/mL and stored at 4°C.
Animals and XJR-Containing Serum Preparation
Male BALB/c (nu/nu) mice (4–5 weeks old, 20–24 g) were purchased from Guangdong Provincial Medical Laboratory Animal Center (Guangdong, China). Mice were maintained under sterile specific pathogen-free conditions at 25°C with 55% humidity and 12 h light/dark cycle, and had free access to water and food. After one week of adaptation, mice were randomly divided into four groups and administered different doses of XJR by gavage for four days continuously: high-dose (HD, 20 g/kg) group, medium-dose (MD, 10 g/kg) group, low-dose (LD, 5 g/kg) group, and blank group (normal saline). Two hours after the last administration of XJR, mice were anesthetized using sodium pentobarbital (50 mg/kg), and the blood in the common carotid artery was collected. The serum (XJR-containing serum) was separated by centrifugation and filtrated through a 0.22 μm filter.
Cell Culture and Treatment
The human normal liver cell line LO2 and the HCC cell lines, HepG2, MHCC97H, Huh7, Bel7402, MHCC97L, and Hep3B (ATCC, Manassas, VA, USA) were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS; HyClone, USA), 100 U/mL penicillin (Gibco, USA), and 100 mg/mL streptomycin (Gibco) at 37°C with 5% CO2. The medium was replaced every two days and cells were used for further experiments after reaching over 80% confluency. To explore the effect of XJR on HCC cells, cells were incubated with XJR-containing serum.
Cell Transfection
miR-29a mimics, miR-29a inhibitor, mimics NC, inhibitor NC, small interfering RNAs (siRNAs) against STAT3 (siSTAT3-1 and −2), and si-NC were all obtained from GenePharma (Shanghai, China). Cell transfection was performed using Lipofectamine 3000 (L3000015; Thermo Fisher Scientific) according to the manufacturer’s protocol.
Luciferase Reporter Gene Assay
TargetScan (http://www.targetscan.org/) was utilized to predict the binding sites between miR-29a and STAT3. The fragment carrying the binding site was cloned into pGL3 vector (Promega, Madison, WI, USA) to generate a STAT3 wild-type (WT) construct. Similarly, the fragment with the mutant binding site was cloned into pGL3 vector to generate a STAT3 mutant (MUT) construct. HCC cells were then co-transfected with miR-29a mimics or mimics NC and STAT3 WT or MUT using Lipofectamine 3000, according to the manufacturer’s instructions. At 48 h post transfection, relative luciferase activity was measured using the luciferase reporter assay system (YPHBio, Beijing, China).
RNA Pull-Down Assay
miR-29a (Bio-miR-29a-WT) and miR-29a mutant (Bio-miR-29a-MUT) were biotinylated using Biotin RNA Labeling Mix (Roche, Basel, Switzerland) and T7/SP6 RNA polymerase (Roche). The cell lysate was incubated with Bio-miR-29a-WT or Bio-miR-29a-MUT for 1 h, followed by incubation with Streptavidin agarose beads (Invitrogen, Carlsbad, CA, USA) for another 1 h. The eluants were then subjected to quantitative real-time polymerase chain reaction (qRT-PCR).
qRT-PCR
Total RNA was extracted from cells using the TRIzol Plus RNA Purification kit (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturers’ protocol. Reverse transcription was performed using the PrimeScript RT Reagent kit (Takara, Dalian, China) and qRT-PCR was performed with SYBR Green I Master (Roche) on the ABI7500 system. The qRT-PCR conditions were as follows: 95°C for 3 min, and 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 20 s. β-actin and U6 were employed as internal controls for STAT3 and miR-29a, respectively. The primer sequences are listed in Table 2. The relative expression level of specific genes was calculated by the 2–ΔΔCt method.
 |
Table 2 The Sequences of Specific Primers |
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
Cells were seeded into 6-well plates at a density of 5 × 103 cells/well. After culturing for 24, 48, and 72 h, cells were incubated with 10 μL MTT solution (5 mg/mL; Sigma-Aldrich, St. Louis, MO, USA) for 3 h at 37°C. DMSO (100 μL) was used to terminate the reaction. The optical density (OD) value at 490 nm wavelength was measured using a microplate reader. A cell proliferation curve was plotted based on the OD values at each time point.
Wound-Healing Assay
Cell migration was detected by the wound-healing assay. Briefly, cells were seeded into 6-well plates and cultured until 80% confluency. Scratches were then made on the cell monolayer using 200 μL pipette tips. The migration distance was measured using an inverted microscope (IX71; Olympus, Japan) at 0 and 24 h post scratching.
Transwell Assay
A transwell chamber (8.0 µm pore size; Costar) was applied to detect the invasion of HCC cells. The surface of the upper chamber was coated with Matrigel (Corning). RPMI-1640 medium (500 μL) was placed in the upper chamber and RPMI-1640 medium (500 μL) containing 10% FBS was added into the lower chamber. The cell suspension (5 × 104 cells/mL) was then inoculated into the upper chamber and cultured for 48 h at 37°C with 5% CO2. Cells on the upper chamber were wiped out with cotton swabs, and cells in the lower chamber were fixed in 4% paraformaldehyde and stained with 0.5% crystal violet. The number of invaded cells were counted in five random fields under a microscope.
Western Blotting
Total protein was isolated from cells using radioimmunoprecipitation assay buffer and quantified using the bicinchoninic acid method. Equal amounts of proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gels and then transferred onto a polyvinylidene fluoride (PVDF) membrane. Subsequently, the PVDF membrane was blocked with 5% fat-free milk for 1 h at 25°C and incubated with specific primary antibodies against STAT3 (1:1000; SAB4502078; Sigma-Aldrich), matrix metalloproteinase (MMP)-2 (1:1000; 40,994; Cell Signaling Technology), MMP-9 (1:1000; 13,667; Cell Signaling Technology), and β-actin (1:1000; 3700; Cell Signaling Technology) at 4°C overnight. After washing three times with TBST for 10 min each, the membrane was incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG secondary antibody (1:5000; 14708s; Cell Signaling Technology) for 1 h at 25°C. The protein bands were visualized using enhanced chemiluminescence reagent and the gray value was determined using an imaging system (Bio-Rad).
Establishment of a Xenograft Tumor Mouse Model
SK-Hep-1 cells subcultured 3–4 times were trypsinized and suspended in serum-free medium. The cell suspension (1.0 × 107 cells/mL) was then subcutaneously injected into the right flank of mice. When the tumor volume reached 70–80 mm3, mice were given different doses of XJR by gavage. The tumor volume was measured every three days for three weeks, and calculated using the following formula: V (mm3) = (a×b2)/2 (a represents the longest diameter and b represents the shortest diameter). After the last measurement, mice were sacrificed by cervical dislocation, and the tumor xenograft was removed and weighed. The inhibition rate (%) was calculated as follows: (1 – tumor weight of treatment group/tumor weight of control group) × 100%. All animal experiments were performed at the animal experiment center of Kunming Municipal Hospital of Traditional Chinese Medicine, and were approved by the Institutional Animal Care and Use Ethics Committee.
Statistical Analysis
All data were presented as mean ± standard deviation (SD) and were analyzed using SPSS 23.0 (SPSS, Chicago, IL, USA) and GraphPad Prism 7.0 (GraphPad Software, San Diego, CA, USA). Comparisons between two groups were made by t-test. Differences among multiple groups were determined by one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparisons test. Pearson’s correlation analysis was utilized to examine the relationship between miR-29a and STAT3 expression in HCC. p < 0.05 was considered statistically significant. Each experiment was performed in triplicate.
Results
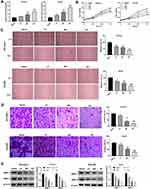
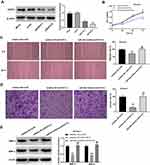
miR-29a Expression Was Significantly Decreased in HCC Tissues and Cells
The expression of miR-29a was significantly lower in HCC tissues than in normal tissues (Figure 1A). The expression of miR-29a was significantly lower in tumors with lymph node (LN)-metastasis than in tumors without LN-metastasis (Figure 1B), as well as in tumors at III–IV stage than in tumors at I–II stage (Figure 1C). In addition, the expression of miR-29a was significantly decreased in HCC cell lines compared with that in human normal liver cell line LO2 (Figure 1D). These results indicated that miR-29a might act as a tumor suppressor in HCC.
XJR Treatment Elevated miR-29a Expression, and Inhibited the Viability, Migration, and Invasion of HCC Cells
In order to explore the effects of XJR on the expression of miR-29a and the malignant behavior of HCC cells, SK-Hep-1 and Hep3B cells were treated with different concentrations of XJR. As shown in Figure 2A, qRT-PCR analysis demonstrated that XJR treatment increased the expression of miR-29a in a concentration-dependent manner. In addition, MTT assay revealed that XJR treatment significantly inhibited cell viability in a dose-dependent manner (Figure 2B). Compared with that in the blank group, the migration of HCC cells was suppressed in the LD, MD, and HD groups, and the migration rate was the lowest in the HD group (Figure 2C). Consistently, the transwell invasion assay showed that the invasion of HCC cells was significantly alleviated by XJR treatment in a dose-dependent manner (Figure 2D). In addition, the protein expression of MMP-2 and −9 was significantly decreased by XJR treatment, especially in the HD group (Figure 2E). These findings indicated that XJR promoted miR-29a expression and HCC development.
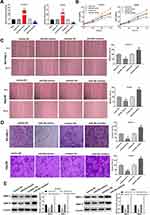
The Regulatory Role of miR-29a in HCC Cell Viability, Migration, and Invasion
Next, to assess the biological function of miR-29a in HCC cells, SK-Hep-1 and Hep3B cells were transfected with miR-29a mimics and inhibitor. qRT-PCR analysis showed that miR-29a expression was remarkably increased and decreased after transfection with miR-29a mimics and inhibitor, respectively (Figure 3A). Furthermore, cell viability was significantly decreased in the miR-29a mimics group compared with that in the mimics NC group, but significantly increased in the miR-29a inhibitor group compared with that in the inhibitor NC group (Figure 3B). Wound-healing and transwell assays showed that cell migration and invasion were significantly decreased by transfection with miR-29a mimics, but significantly increased by transfection with miR-29a inhibitor (Figure 3C and D). In addition, the protein expression of MMP-2 and −9 was significantly decreased by transfection with miR-29a mimics, but increased by transfection with miR-29a inhibitor (Figure 3E). Collectively, these data suggested that miR-29a suppressed the viability, migration, and invasion of HCC cells.
Down-Regulation of miR-29a Reversed the XJR-Induced Inhibitory Effects on Cell Malignant Behaviors
To investigate the potential correlation between XJR and miR-29a in HCC progression, functional experiments were performed in SK-Hep-1 cells. As shown in Figure 4A, the increase in miR-29a expression in the HD treatment group was significantly inhibited by transfection with miR-29a inhibitor. Similarly, the XJR-induced inhibitory effects on the viability, migration, and invasion of SK-Hep-1 cells in the HD group were significantly reversed by transfection with miR-29a inhibitor (Figure 4B–D). In addition, down-regulation of miR-29a significantly reversed the XJR-induced increase in MMP-2 and −9 protein expression in the HD group (Figure 4E). Taken together, our data suggest that down-regulation of miR-29a can reverse the anti-tumor effect of HD XJR on SK-Hep-1 cells.
STAT3 Was a Target of miR-29a
A direct binding site between miR-29a and STAT3 was predicated by TargetScan (Figure 5A). In order to identify the correlation between miR-29a and STAT3, luciferase reporter gene and RNA pull-down assays were performed. The luciferase reporter gene assay showed that luciferase activity was significantly lower in cells co-transfected with STAT3-WT and miR-29a mimics than in cells co-transfected with STAT3-WT and NC-mimics (Figure 5B). RNA pull-down assay showed that STAT3 expression was higher in the Bio-miR-29a-WT group than in the Bio-NC group (Figure 5C). In addition, STAT3 expression was found to be significantly higher in HCC tissues than in normal tissues (Figure 5D). Moreover, STAT3 expression was significantly higher in HCC cells than in LO2 cells (Figure 5E). Linear regression analysis showed that STAT3 expression was negatively associated with miR-29a expression in HCC tissues (Figure 5F). In summary, STAT3 was identified as a direct target of miR-29a in HCC.
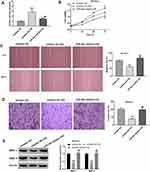
Down-Regulation of miR-29a Reversed the Suppressive Effect of Si-STAT3 on HCC Cells
Because STAT3 was identified as a direct target of miR-29a, we assumed that treatment with miR-29a inhibitor would inhibit HCC progression through negatively regulating STAT3. To validate this hypothesis, SK-Hep-1 cells were transfected with siSTAT3-1 and siSTAT3-2 to down-regulate STAT3 expression. Western blot analysis revealed that the protein expression of STAT3 was significantly decreased in SK-Hep-1 cells by transfection with siSTAT3-1 and siSTAT3-2 (Figure 6A). siSTAT3-2 with relatively high silencing efficiency was used for further experiments. Transfection with siSTAT3-2 significantly inhibited the viability, migration, and invasion of SK-Hep-1 cells; however, transfection with miR-29a inhibitor significantly reversed these effects (Figure 6B–D). In addition, the protein expression of MMP-2 and −9 was significantly decreased by transfection with siSTAT3-2, but this effect was attenuated by transfection with miR-29a inhibitor (Figure 6E). These results indicated that the inhibitory effects of siSTAT3-2 on cell viability, migration, and invasion were reversed by miR-29a inhibitor.
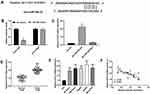
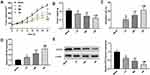
XJR Inhibited Tumor Growth in Mice
The effects of different concentrations of XJR on HCC development in vivo were analyzed in mice. Results showed that tumor volume and weight were significantly decreased by XJR treatment in a dose-dependent manner (Figure 7A and B). Besides, the tumor inhibition rate was significantly increased with increasing concentrations of XJR (Figure 7C). In addition, the expression of miR-29a was significantly increased, and the protein expression of STAT3 was significantly decreased by treatment with XJR in a dose-dependent manner (Figure 7D and E). These results indicated that XJR suppressed tumor growth in mice via regulating miR-29a and STAT3.
Discussion
HCC, the most common type of liver cancer, is a public health risk due to its high occurrence rate and unfavorable outcomes.26 Most cases of HCC have been reported in developing countries, and China is the most high-risk region of HCC in the world.27 TCM exerts anti-tumor effects at multiple levels through systematic regulation of the body, such as suppressing and killing tumor cells, regulating immune function, improving symptoms and signs, and reducing radiochemotherapy-related toxicity.11 XJR, identified by Professor Zhongying Zhou at Nanjing University of Traditional Chinese Medicine, exhibits satisfactory anti-tumor effect and has been used in clinical practice in China for many years. In this study, XJR-containing serum was prepared to simulate the environment of drug intervention in vivo. The inhibitory effects of XJR on HCC were then evaluated in HCC cells and mouse model. In vitro experiments showed that XJR treatment inhibited the viability, migration, and invasion of HCC cells. The anti-tumor effect of XJR was gradually increased with increasing concentration. In vivo experiments showed that XJR treatment significantly suppressed tumor growth in mice in a dose-dependent manner. These findings illustrate the anti-tumor role of XJR in HCC. However, some active ingredients in the serum may affect the efficacy evaluation. Furthermore, it is necessary to determine the effective drug concentration in the serum to eliminate the complexity of serum administration.
In this study, XJR-treated HCC cells displayed increased expression of miR-29a. Previously, Wang et al demonstrated that miR-29a inhibited the proliferation and migration of HCC cells through down-regulating the expression of IGF1R.28 In addition, Kogure et al showed that miR-29a is involved in the epigenetic regulation of transforming growth factor-β-induced epithelial-to-mesenchymal transition in HCC.29 Here, to verify the regulatory role of miR-29a in HCC, the expression of miR-29a was detected in 40 patients with HCC and 40 healthy volunteers. The results showed that miR-29a expression was significantly decreased in HCC tissues. Moreover, down-regulation of miR-29a was associated with LN metastasis and tumor node metastasis (TNM) stages. As expected, overexpression of miR-29a inhibited the proliferation, migration, and invasion of HCC cells, while silencing of miR-29a promoted the cell malignant behaviors. These data are consistent with previous data on the anti-cancer effects of miR-29a in HCC.30–32 Furthermore, silencing of miR-29a reversed the inhibitory effects of HD XJR on the proliferation, migration, and invasion of HCC cells. These results indicate that XJR may inhibit HCC tumorigenesis through up-regulation of miR-29a.
The regulatory effect of miRNAs on tumorigenesis is usually mediated by interacting with target genes. In this study, the potential targets of miR-29a were predicated using TargetScan. STAT3 was identified as a direct target of miR-29a in HCC. STAT3, an important transcription factor, plays a vital role in tumorigenesis.33,34 Numerous studies have confirmed that STAT3 can induce cancer development via DNA methylation and modulation of chromatin topology.35 Over the past decades, STAT3 has been widely used as a therapeutic target for HCC treatment. For instance, STAT3-blocked whole-cell hepatoma vaccine has been shown to elicit cellular and humoral immune responses against HCC.36 Moreover, STAT3 was shown to markedly aggravate epithelial-to-mesenchymal transition and migration in HCC cells.37 Additionally, STAT3 mediates HCC metastasis through regulating HOXD-AS1.38 In this study, luciferase reporter gene and RNA pull-down assays validated the interaction between STAT3 and miR-29a in HCC cells. Pearson’s correlation analysis showed that STAT3 expression was negatively correlated with miR-29a expression. In addition, down-regulation of STAT3 inhibited HCC cell viability, migration, and invasion; however, these anti-tumor effects were reversed by treatment with miR-29a inhibitor. These results indicate that miR-29a may inhibit HCC tumorigenesis by down-regulating the expression of STAT3.
Conclusions
In conclusion, XJR treatment effectively inhibited HCC cell proliferation, migration, and invasion in vitro and suppressed tumor growth in vivo through regulating miR-29a and STAT3 expression. These data suggest that XJR may serve as a promising drug for the treatment of HCC. Furthermore, our findings provide insights into the anti-tumor role of XJR in HCC as well as its underlying mechanism of action.
Abbreviations
HCC, Hepatocellular carcinoma; XJR, Xiaoai Jiedu Recipe; qRT-PCR, Quantitative real-time polymerase chain reaction; LN, Lymph node; TCM, Traditional Chinese medicine; STAT3, Signal transducer and activator of transcription 3; OD, Optical density.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Informed Consent
This study was approved by the Ethics Committee of Kunming Municipal Hospital of Traditional Chinese Medicine in accordance with the Declaration of Helsinki. Written informed consent was obtained from all patients. All animal experiments were performed in the animal experiment center of Kunming Municipal Hospital of Traditional Chinese Medicine, and were approved by the Institutional Animal Care and Use Ethics Committee. All participants agreed the study could be published.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Fan J-H, Wang J-B, Jiang Y, et al. Attributable causes of liver cancer mortality and incidence in china. Asian Pac j Cancer Preve. 2013;14(12):7251–7256. doi:10.7314/APJCP.2013.14.12.7251
3. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/S0140-6736(18)30010-2
4. Liang S, Zou Y, Gao J, et al. The Chinese medicine, Jiedu Recipe, inhibits the epithelial mesenchymal transition of hepatocellular carcinoma via the regulation of smad2/3 dependent and independent pathways. Evidence Based Complementary Altern Med. 2018;2018:5629304. doi:10.1155/2018/5629304
5. Medavaram S, Zhang Y. Emerging therapies in advanced hepatocellular carcinoma. Exp Hematol Oncol. 2018;7(1):17. doi:10.1186/s40164-018-0109-6
6. Xin P, Kuang H-X, Li X-L, et al. Proteomics and its application to determine mechanism of action of traditional Chinese medicine. Zhongguo Zhong Yao Za Zhi. 2018;43(5):904–912. doi:10.19540/j.cnki.cjcmm.20180118.002
7. Olugbami JO, Damoiseaux R, France B, et al. Atomic force microscopy correlates antimetastatic potentials of HepG2 cell line with its redox/energy status: effects of curcumin and Khaya senegalensis. J Integr Med. 2017;15(3):214–230. doi:10.1016/S2095-4964(17)60337-6
8. Ruan S-M, Shen M-H, Lin H, et al. Jiedu sangen decoction intervened carcinoma-associated fibroblasts and inhibited migration and invasion of colon cancer: an experimental research. Zhongguo Zhongxiyi Jiehe Zazhi. 2013;33(9):1242–1246.
9. Yuan L, Zhou M, Wasan HS, et al. Jiedu sangen decoction inhibits the invasion and metastasis of colorectal cancer cells by regulating EMT through the hippo signaling pathway. Evidence Based Complementary Altern Med. 2019;2019:1431726. doi:10.1155/2019/1431726
10. Yu Y, Lang Q, Chen Z, et al. The efficacy for unresectable hepatocellular carcinoma may be improved by transcatheter arterial chemoembolization in combination with a traditional Chinese herbal medicine formula: a retrospective study. Cancer. 2009;115(22):5132–5138. doi:10.1002/cncr.24567
11. Chen LY, Zhai XF, Chen Z, et al. Jie-du granule preparation for the treatment of advanced hepatocellular carcinoma: a retrospective cohort study of 177 patients. Oncotarget. 2017;8(18):30471–30476. doi:10.18632/oncotarget.12887
12. Zhou HG, Chen HB, Mian-Hua WU, et al. Clinical observation of Xiao’ai Jiedu prescription combined with chemotherapy on advanced cancer. Chin J Traditional Chin Med Pharm. 2010;25(7):1140–1143.
13. Shi WJ, Tan JN, Shen WX, Chang-Liang XU, Sun DD, Cheng HB. Discussion on inhibitory effect and mechanism of Xiaoai Jiedu recipe on colon cancer. Chin J Traditional Chin Med Pharm. 2018;33(1):69–72.
14. Chen HB, Shen B, Li LI, et al. Xiaoai Jiedu recipe inhibits transplanted hepatocarcinoma H22 tumors and the related mechanism. Chin J Cancer Biother. 2011;18(1):28–32.
15. Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57(2):840–847. doi:10.1002/hep.26095
16. Xue X, Zhao Y, Wang X, et al. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J Cell Biochem. 2019;120(1):135–142. doi:10.1002/jcb.27165
17. Mahati S, Xiao L, Yang Y, et al. miR-29a suppresses growth and migration of hepatocellular carcinoma by regulating CLDN1. Biochem Biophys Res Commun. 2017;486(3):732–737. doi:10.1016/j.bbrc.2017.03.110
18. Zhang Y, Yang L, Wang S, Liu Z, Xiu M. MiR-29a suppresses cell proliferation by targeting SIRT1 in hepatocellular carcinoma. Cancer Biomark. 2018;22(1):151–159. doi:10.3233/CBM-171120
19. Qiu FL, Chen HB, Jiang ZQ, Zhou HG. Effect of Xiaoai Jiedu recipe on miRNA expression profiles in H22 tumor-bearing mice. Chin J Integr Traditional West Med. 2016;36(9):1112–1118.
20. Barton BE. STAT3: a molecular target for cancer whose time has come. Curr Signal Transduct Ther. 2016;1(3):239–253. doi:10.2174/157436206778226978
21. Svinka J, Mikulits W, Eferl R. STAT3 in hepatocellular carcinoma: new perspectives. Hepatic Oncol. 2014;1(1):107–120. doi:10.2217/hep.13.7
22. Zuo M, Li C, Lin J, Javle M. LLL12, a novel small inhibitor targeting STAT3 for hepatocellular carcinoma therapy. Oncotarget. 2015;6(13):10940–10949. doi:10.18632/oncotarget.3458
23. Jung KH, Yoo W, Stevenson HL, et al. Multifunctional effects of a small-molecule STAT3 inhibitor on NASH and hepatocellular carcinoma in mice. Clin Cancer Res. 2017;23(18):5537–5546. doi:10.1158/1078-0432.CCR-16-2253
24. Liu S, Zhang X, Hu C, Wang Y, Xu C. miR-29a inhibits human retinoblastoma progression by targeting STAT3. Oncol Rep. 2018;39(2):739–746. doi:10.3892/or.2017.6144
25. Gao J, Shao Z, Yan M, Fu T, Zhang L, Yan Y. Targeted regulation of STAT3 by miR-29a in mediating Taxol resistance of nasopharyngeal carcinoma cell line CNE-1. Cancer Biomark. 2018;22(4):641–648. doi:10.3233/CBM-170964
26. Lin Y, Jian Z, Jin H, et al. Long non-coding RNA DLGAP1-AS1 facilitates tumorigenesis and epithelial-mesenchymal transition in hepatocellular carcinoma via the feedback loop of miR-26a/b-5p/IL-6/JAK2/STAT3 and Wnt/β-catenin pathway. Cell Death Dis. 2020;11(1):34. doi:10.1038/s41419-019-2188-7
27. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int j Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
28. Wang X, Liu S, Cao L, et al. miR-29a-3p suppresses cell proliferation and migration by downregulating IGF1R in hepatocellular carcinoma. Oncotarget. 2017;8(49):86592–86603. doi:10.18632/oncotarget.21246
29. Kogure T, Kondo Y, Kakazu E, et al. Involvement of miRNA-29a in epigenetic regulation of transforming growth factor-β-induced epithelial-mesenchymal transition in hepatocellular carcinoma. Hepatol Res. 2014;44(8):907–919. doi:10.1111/hepr.12188
30. Liang Y, Li E, Min J, et al. miR-29a suppresses the growth and metastasis of hepatocellular carcinoma through IFITM3. Oncol Rep. 2018;40(6):3261–3272. doi:10.3892/or.2018.6745
31. Zhu X-C, Dong Q-Z, Zhang X-F, et al. microRNA-29a suppresses cell proliferation by targeting SPARC in hepatocellular carcinoma. Int J Mol Med. 2012;30(6):1321–1326. doi:10.3892/ijmm.2012.1140
32. Xiao Z, Wang Y, Ding H. XPD suppresses cell proliferation and migration via miR-29a-3p-Mdm2/PDGF-B axis in HCC. Cell Biosci. 2019;9(1):6. doi:10.1186/s13578-018-0269-4
33. Galoczova M, Coates P, Vojtesek B. STAT3, stem cells, cancer stem cells and p63. Cell Mol Biol Lett. 2018;23(1):12. doi:10.1186/s11658-018-0078-0
34. Fathi N, Rashidi G, Khodadadi A, et al. STAT3 and apoptosis challenges in cancer. Int J Biol Macromol. 2018;117:993–1001. doi:10.1016/j.ijbiomac.2018.05.121
35. Yu H, Lee H, Herrmann A, et al. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nature Reviews Cancer. 2014;14(11):736–746. doi:10.1038/nrc3818
36. Han Q, Wang Y, Pang M, et al. STAT3-blocked whole-cell hepatoma vaccine induces cellular and humoral immune response against HCC. J Exp Clin Cancer Res. 2017;36(1):156. doi:10.1186/s13046-017-0623-0
37. Wang B, Liu T, Wu J-C, et al. STAT3 aggravates TGF-β1-induced hepatic epithelial-to-mesenchymal transition and migration. Biomed Pharmacother. 2018;98:214–221. doi:10.1016/j.biopha.2017.12.035
38. Wang H, Huo X, Yang X-R, et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol Cancer. 2017;16(1):136. doi:10.1186/s12943-017-0680-1
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.