Back to Journals » OncoTargets and Therapy » Volume 12
TNF-like ligand 1A is associated with progression and prognosis of human gastric cancer
Authors Gao Y, Wang Y, Wang X , Wang Y, Zhang X, Sun X
Received 2 April 2019
Accepted for publication 13 August 2019
Published 19 September 2019 Volume 2019:12 Pages 7715—7723
DOI https://doi.org/10.2147/OTT.S210939
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yaxian Gao,1,2,* Yuanyuan Wang,3,* Xiao Wang,1 Yongwei Wang,4 Xiaoqing Zhang,1 Xun Sun1
1Department of Immunology, China Medical University, Shenyang, Liaoning 110000, People’s Republic of China; 2Department of Immunology, Chengde Medical College, Chengde, Hebei 067000, People’s Republic of China; 3Department of Anesthesiology, The Fourth Affiliated Hospital, China Medical University, Shenyang, Liaoning 110000, People’s Republic of China; 4Department of Anatomy, Chengde Medical College, Chengde, Hebei 067000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xun Sun
Department of Immunology, China Medical University, No. 77 Puhe Road, North New Area, Shenyang, Liaoning 110000, People’s Republic of China
Tel +86 242 325 6666 (ext. 5345)
Fax +86 242 325 5291
Email [email protected]
Purpose: This study aimed to investigate the function of TNF-like ligand 1A (TL1A) in the tumorigenesis and progression of gastric cancer (GC).
Methods: RNA-seq gene expression and clinical information for GC patients were obtained from The Cancer Genome Atlas (TCGA) database. Differentially expressed genes (DEGs) between GC tissue samples and normal controls were screened with the edgeR package. Identification of gene co-expression and functional enrichment analyses were performed with Pearson’s correlation analysis and gene set enrichment analysis (GSEA), respectively. Lastly, survival analysis was performed using the Kaplan-Meier method with the log rank test.
Results: TL1A expression in GC tissue samples were significantly higher than that in normal controls (LogFC=1.07 and P=8.90E-07). Moreover, 215 genes, co-expressed with TL1A, and 21 Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were obtained. Next, the miRNA-lncRNA/mRNA network, comprising 7 miRNAs, 27 lncRNAs, and 21 mRNAs, was constructed based on key genes from intersections between co-expression analysis and GSEA. In addition, survival analysis results demonstrated that TL1A (P=2.6e−07) was significantly associated with the overall survival (OS) of GC patients.
Conclusion: TL1A was involved in the tumorigenesis and progression of GC, and was significantly associated with the OS of GC patients.
Keywords: TNF-like ligand 1A, gastric cancer, co-expression analysis, GSEA, prognosis
Introduction
Gastric cancer (GC) is the fifth most common malignancy, and the third leading cause of cancer-related deaths (723,100 in 2012) globally.1 Currently, a series of risk factors, such as dietary factors, Epstein-Barr virus, and Helicobacter pylori infection have been identified for GC. Moreover, an increasing amount of evidence show that genetic factors and their downstream effects on cellular processes play important roles in GC progression, but, the pathogenesis of GC still remains complex.2,3 Additionally, despite the advances in cancer diagnosis and treatment, the prognosis of GC patients still remains unsatisfactory with the 5-year survival rate less than 30% due to its aggressive behavior.4 Hence, it is urgent to identify novel biomarkers and effective therapeutic targets for patients with GC.
TNF-like ligand 1A (TL1A; also known as TNFSF15 or VEGI), is a unique cytokine produced by endothelial cells, T cells, macrophages, monocytes and dendritic cells, which inhibits proliferation and angiogenesis of endothelial cells.5,6 TL1A has been reported to be involved in modulating vascular homeostasis and inflammation.7 Previous studies have reported that TL1A plays an important role in a variety of autoimmune inflammatory diseases, such as rheumatic arthritis (RA), asthma and inflammatory bowel disease.8 Additionally, TL1A is involved in the development of multiple tumor types,9,10 and is recognized as the potential therapeutic target for cancer therapy.11,12 Recently, reduced TL1A expression was found to be associated with poor prognosis in breast cancer patients.11 However, the role of TL1A in GC progression remains unclear.
In the present study, in order to reveal the potential role of TL1A in tumorigenesis of GC, we screened the differentially expressed genes (DEGs) and co-expressed genes associated with TL1A in GC. Subsequently, we performed gene set enrichment analysis (GSEA) to obtain the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. Finally, we constructed a TL1A-related miRNA-lncRNA/mRNA network and predicted the prognostic value of TL1A in GC.
Materials and methods
Data collection
Clinical and survival data from 418 patients with GC were retrieved from The Cancer Genome Atlas (TCGA) through the Genomic Data Commons (GDC) portal. Further, 375 tumor samples and 50 normal tissues samples with the RNA-seq gene expression data (HTSeq–Counts) were included in the present study. All data in our study were downloaded from the database of University of California Santa Cruz (UCSC) xena (https://xenabrowser.net).
Data preprocessing and differential expression analysis
Since the downloaded RNA-seq expression profile data were converted logarithmically, we first transformed it into a read count generated by normal HTSeq software.13 In addition, the edgeR package14 was used to screen the DEGs between GC tissues samples and normal controls in the expressing data. Next, information on the obtained genes was supplemented by combining the gene annotation information in GENCODE database.15
Identification of co-expression gene of TL1A and GSEA
Pearson’s correlation matrices were performed for the analysis of the correlation between TL1A and other genes. Genes with correlation coefficients >0.5 (P<0.01) were considered to have co-expressed relationship with TL1A. Moreover, according to gene-type information, lncRNA was selected as the key gene for downstream analysis.
To obtain the KEGG pathway associated with TL1A, we conducted GSEA, as previously described,16 based on the background of all the genes. Results with ES >0.5, NES >1.7 and P-value <0.05 were regarded as significant. Gene sets derived from the enrichment analysis were integrated with mRNA from the co-expression analysis to identify overlapped genes as key genes.
Construction of the tl1a-related miRNA-lncRNA/mRNA network
In order to construct the miRNA-lncRNA/mRNA network based on the relationship between miRNA and lncRNA or mRNA, we downloaded the lncRNA transcript sequence, mRNA sequence of 3ʹUTR and mature miRNA sequences from the Ensembl database to predict the relationship between miRNA and lncRNA or mRNA using miRanda.17 Furthermore, the interactions were in accordance with the following criteria: 1) miRNA had a regulatory relationship with TL1A; 2) miRNA simultaneously regulated both mRNA and lncRNA; and 3) miRNA regulated more than 10 mRNAs or lncRNAs. Next, Cytoscape was used to construct and visualize the miRNA-lncRNA/mRNA regulatory network.18
Validation of DEGs based on clinical samples and cell lines
For further validation of our findings, TL1A expression levels were also determined in clinical 10 GC tissues samples and corresponding adjacent normal samples from the Affiliated Hospital of Chengde Medical College, as well as normal human stomach cells GES-1 and a panel of GC cells (AGS, BGC823, MGC803, SGC7901, and HGC27), which were purchased from the Chinese Academy of Sciences, Shanghai, China. This study was approved by the Ethical Committee of Affiliated Hospital of Chengde Medical College, and all patients signed informed consents.
Total RNA from tissues and cells were extracted using Trizol reagent (Invitrogen). First-strand cDNA was generated using Reverse Transcription Reagents (Takara RR047) according to the manufacturer’s protocol. Real-time qPCR was performed in the Real-Time PCR Detection System (Bio-Rad) using SYBR Green (Takara RR820). Primer sequences were as follows: TL1A, F: 5′-TTAGAGCAGACGGAGATAAGCCAAG-3′; R: 5′-CCTAGTTCATGTTCCCAGTGCAGA-3′ and β-actin, F: 5′-AGCGAGCATCCCCCAAAGTT-3′; R: 5′-GGGCACGAAGGCTCATCATT-3′. Relative expression of TL1A normalized to β-actin was calculated with the 2–ΔΔCt method.
Survival analysis
To evaluate the prognostic value of the screened genes, survival analysis was performed, and to improve the reliability of the results with larger samples, we performed a KM plot analysis (http://kmplot.com) using a JetSet best probe set (Only including HGU133 PLUS 2.0 microarray data), based on the datasets, GSE14210, GSE15459, GSE22377, GSE29272, GSE51105, and GSE62254. A P<0.05 was considered statistically significant.
Results
Expression pattern of TL1A in GC
As shown in Figure 1, multidimensional scaling (MDS) analysis of the samples revealed that the tumor samples were concentrated in the middle of the image in compared to the normal controls. Moreover, TL1A expression in GC samples was higher than that in normal controls (Log FC =1.07), and the difference was significant (P=8.90E-07; Table 1).
 |
Table 1 The information of TL1A |
 |
Figure 1 Multidimensional scaling (MDS) analysis of samples. Compared to normal tissue samples (green circle), tumor samples (red square) were concentrated in the center of the image. |
Co-expression analysis
A total of 215 genes co-expressed with TL1A were obtained after Pearson’s correlation analysis. As illustrated in Table 2, the top ten genes more closely correlated with TL1A, including caspase 10 (CASP10), iodotyrosine deiodinase (IYD), major vault protein (MVP), g protein-coupled receptor class c group 5 member A (GPRC5A), solute carrier family 45 member 4 (SLC45A4), interferon gamma receptor 1 (IFNGR1), rho guanine nucleotide exchange factor 5 (ARHGEF5), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta (PIK3CB), lymphocyte antigen 75 (LY75), and caspase 8 (CASP8) were identified (all, P<0.05).
 |
Table 2 Analysis of co-expression genes with TL1A (top 10) |
Functional prediction based on GSEA
We performed GSEA to identify the potential function of the screened DEGs. A total of 21 KEGG pathways were enriched (ES >0.5, NES >1.7, P<0.05; Table 3). Furthermore, based on the enriched gene sets and the genes from the co-expression analysis, we identified the intersection genes as the key genes that are relevant to the disease progression of GC. Moreover, the relationships between the intersection genes and the enriched KEGG pathway were visualized as interaction networks (Figure 2).
 |
Table 3 KEGG pathways significantly enriched from GSEA |
Construction of miRNA-lncRNA/mRNA network
Based on the above data, the miRNA-lncRNA/mRNA network was constructed using Cytoscape. As illustrated in Figure 3, the miRNA-lncRNA/mRNA network was composed of 7 miRNA nodes, 27 lncRNA nodes, and 21 mRNA nodes.
 |
Figure 3 The miRNA-lncRNA/mRNA network in GC. Blue triangles represent miRNA. Orange diamonds represent lncRNA. Circles represent mRNA. |
Identification of survival-related lncRNAs in GC
In order to investigate the genes associated with the survival of GC patients, 876 patients were divided into the high and low groups based on the gene expression median. The results showed that the patients with high TL1A levels had shorter overall survival (OS) than those with low expression (P=2.6e−07; Figure 4A). Similarly, SRC Proto-Oncogenes (SRC) were positively correlated with the OS of GC patients, respectively (P=0.0081; Figure 4B).
 |
Figure 4 Survival analysis using the Kaplan-Meier curve. Patients with high expression of TL1A (A) and SRC (B) had worse OS than those with low expression. |
Validation of TL1A in clinical tissue samples and cell lines
To confirm the reliability of the TL1A identified in silico, the TL1A expression profiles were verified in tissue samples and cell lines. TL1A levels in ten GC tissues were all higher than those in adjacent normal samples (Figure 5), which is in accordance with the data above. Similarly, TL1A expression in GC cell lines were all significantly upregulated compared to that in normal cell lines (all, P<0.05).
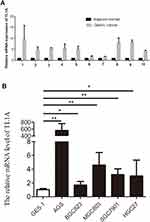 |
Figure 5 TL1A expression is determined by qRT-PCR. The TL1A mRNA levels in GC tissues (A) and cell lines (B) were higher than those in normal tissue samples and cell lines. *P<0.05 and **P<0.01. |
Discussion
Despite developments in GC treatment, its prognosis remains frustratingly poor. Hence, the identification of risk factors and accurate prognostic prediction in GC patients is still urgent. In this study, we determined the clinical significance of TL1A in GC progression based on TCGA database. The results showed that TL1A expression in GC tissues samples was significantly increased compared to that in normal controls. Moreover, a set of co-expressed genes, enriched genes, and KEGG pathways were obtained. Then, the miRNA-lncRNA/mRNA network was constructed that provided an integrated view of RNA regulation. After a series of bioinformatics analyses, TL1A was identified to be associated with the progression and prognosis of GC.
In fact, as a counter-balancing factor in the maintenance of vascular integrity, TL1A has been found to be abnormally expressed in multiple tumor types.19,20 Moreover, Qin et al21 reveals that TL1A could promote lymphatic metastasis of lung cancer cells via VEGFC up-regulation. These studies suggest that TL1A may play an important role in tumorigenesis, and in accordance with them, our results, based on the data from TCGA database, suggest that TL1A expression levels in GC tissue samples increased significantly when compared to normal controls. Furthermore, the validation analysis showed that TL1A expression was upregulated in other clinical GC tissue samples and cell lines. In addition, co-expression analysis detected 215 genes that were co-expressed with TL1A. Among these co-expressed genes, CASP10 is reported to be associated with the risk of GC,22 and GPRC5A overexpression could predict the advanced poor prognosis in patients with GC.23 All these results suggested that TL1A, along with a series of co-expressed genes are involved in the development of GC; however, the mechanism of function still requires further investigation.
Recent studies suggest TL1A can suppress proliferation and induce apoptosis of endothelial cells through different signaling pathways.24,25 Based on KEGG pathway analysis with GSEA, some cancer-related pathways, such as the phosphatidylinositol, NOD-like receptor (NLR), and JAK-STAT signaling pathways, were determined, indicating that TL1A may play an important role in GC development through different pathways. Chiappini et al26 reported the prognostic implications of the phosphatidylinositol signaling pathway activation in GC. Castaño-Rodríguez et al27 highlighted the relationship between the NLR signaling pathway and gastric carcinogenesis. Additionally, the involvement of the JAK-STAT signaling pathway in gastric carcinogenesis has been reported.28,29 Hence, via pathway analysis, we further verified that TL1A might be an important factor in GC development.
Until now, few studies have investigated the interaction between miRNA and lncRNAs, or miRNAs and mRNAs in GC. In this study, we constructed a miRNA-lncRNA/mRNA network with 7 miRNAs, 27 lncRNAs, and 21 mRNAs, which provided an integrated view of the regulatory crosstalk among these genes associated with TL1A in GC. More importantly, TL1A, along with SRC, were all found to be significantly associated with the survival of GC patients. Specifically, patients with high expression levels of TL1A, or SRC had shorter survival times than those with low expression. In fact, accumulating evidence indicates that TL1A was associated with GC prognosis.11,30 SRC, a nonreceptor tyrosine kinase that acts downstream of receptor tyrosine kinases, is involved in cell proliferation migration, adhesion, and invasion.31 Elsberger et al32 reported that high expression levels of cytoplasmic SRC was associated with a worse prognosis of luminal-type breast cancer, which is in accordance with our studies.
Conclusion
In conclusion, we identified that TL1A was aberrantly expressed in GC; meanwhile, a set of genes co-expressed with TL1A, and pathways were detected. Furthermore, we constructed the miRNA-lncRNA/mRNA network, and identified that TL1A was correlated with OS in GC patients. To our knowledge, this is the first study to evaluate the function of TL1A in the tumorigenesis and progression of GC. However, the clinical value of TL1A for targeted GC therapy and prognosis still needs to be verified.
Data sharing statement
All data generated or analyzed during this study are included in this published article.
Acknowledgment
This study was funded by the National Natural Science Foundation of China (grant number 31370921), Research Project for Hebei Provincial College of Science and Technology (grant number QN2015044), and Research Project for Hebei Provincial College of Science and Technology (grant number QN2016060).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflict of interest in this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Gigek CO, Calcagno DQ, Rasmussen LT, et al. Genetic variants in gastric cancer: risks and clinical implications. Exp Mol Pathol. 2017;103(1):101–111. doi:10.1016/j.yexmp.2017.07.004
3. Razzak M. Genetics: new molecular classification of gastric adenocarcinoma proposed by The Cancer Genome Atlas. Nat Rev Clinl Oncol. 2014;11(9):499. doi:10.1038/nrclinonc.2014.138
4. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi:10.3322/caac.21166
5. Cavallini C, Lovato O, Bertolaso A, et al. Expression and function of the TL1A/DR3 axis in chronic lymphocytic leukemia. Oncotarget. 2015;6(31):32061. doi:10.18632/oncotarget.5201
6. Zhai Y, Yu J, Iruela‐Arispe L, et al. Inhibition of angiogenesis and breast cancer xenograft tumor growth by VEGI, a novel cytokine of the TNF superfamily. Int J Cancer. 1999;82(1):131–136. doi:10.1002/(sici)1097-0215(19990702)82:1<131::aid-ijc22>3.0.co;2-o
7. Zhang Z, Li L-Y. TNFSF15 modulates neovascularization and inflammation. Cancer Microenvironment. 2012;5(3):237–247. doi:10.1007/s12307-012-0117-8
8. Levin I, Zaretsky M, Aharoni A. Directed evolution of a soluble human DR3 receptor for the inhibition of TL1A induced cytokine secretion. PLoS One. 2017;12(3):e0173460. doi:10.1371/journal.pone.0173460
9. Zhou J, Yang Z, Tsuji T, et al. LITAF and TNFSF15, two downstream targets of AMPK, exert inhibitory effects on tumor growth. Oncogene. 2011;30(16):1892. doi:10.1038/onc.2010.575
10. Deng W, Gu X, Lu Y, et al. Down-modulation of TNFSF15 in ovarian cancer by VEGF and MCP-1 is a pre-requisite for tumor neovascularization. Angiogenesis. 2012;15(1):71–85. doi:10.1007/s10456-011-9244-y
11. Parr C, Gan CH, Watkins G, Jiang WG. Reduced vascular endothelial growth inhibitor (VEGI) expression is associated with poor prognosis in breast cancer patients. Angiogenesis. 2006;9(2):73–81. doi:10.1007/s10456-006-9033-1
12. Wu J, Jiang Y, Yang W, et al. Dual function of RGD-modified VEGI-192 for breast cancer treatment. Bioconjug Chem. 2012;23(4):796–804. doi:10.1021/bc2006576
13. Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi:10.1093/bioinformatics/btu638
14. Lun AT, Chen Y, Smyth GK. It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. In: Statistical Genomics. Springer; 2016;391–416.
15. Frankish A, Harrow J. GENCODE pseudogenes. In: Pseudogenes. Springer; 2014:129–155.
16. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
17. Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi:10.1186/gb-2003-5-1-r1
18. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
19. Zhang N, Sanders AJ, Ye L, Kynaston HG, Jiang WG. Expression of vascular endothelial growth inhibitor (VEGI) in human urothelial cancer of the bladder and its effects on the adhesion and migration of bladder cancer cells in vitro. Anticancer Res. 2010;30(1):87–95.
20. Chew L-J, Pan H, Yu J, et al. A novel secreted splice variant of vascular endothelial cell growth inhibitor. Faseb J. 2002;16(7):742–744. doi:10.1096/fj.01-0757fje
21. Qin T, Huang D, Liu Z, et al. TNFSF15 promotes lymphatic metastasis via up-regulation of VEGFC in a mouse model of lung cancer. Cancer Sci. 2018. doi:10.1111/cas.13665
22. Hyland PL, Lin SW, Hu N, et al. Genetic variants in fas signaling pathway genes and risk of gastric cancer. Int J Cancer. 2014;134(4):822–831. doi:10.1002/ijc.28415
23. Liu H, Zhang Y, Hao X, et al. GPRC5A overexpression predicted advanced biological behaviors and poor prognosis in patients with gastric cancer. Tumor Biol. 2016;37(1):503–510. doi:10.1007/s13277-015-3817-0
24. Grimaldo S, Tian F, Li L-Y. Sensitization of endothelial cells to VEGI-induced apoptosis by inhibiting the NF-κB pathway. Apoptosis. 2009;14(6):788–795. doi:10.1007/s10495-009-0351-9
25. Haridas V, Shrivastava A, Su J, et al. VEGI, a new member of the TNF family activates nuclear factor-κB and c-Jun N-terminal kinase and modulates cell growth. Oncogene. 1999;18(47):6496. doi:10.1038/sj.onc.1203170
26. Chiappini PBO, De Medeiros IUD, Lima LGC, et al. Prognostic implications of phosphatidylinositol 3-kinase/AKT signaling pathway activation in gastric carcinomas. Arch Med Sci. 2017;13(6):1262. doi:10.5114/aoms.2016.60394
27. Castaño-Rodríguez N, Kaakoush NO, Goh KL, Fock KM, Mitchell HM. The NOD-like receptor signalling pathway in Helicobacter pylori infection and related gastric cancer: a case-control study and gene expression analyses. PLoS ONE. 2015;10(1): e98899. doi: 10.1371/journal.pone.0117870
28. Xiao C, Hong H, Yu H, et al. MiR-340 affects gastric cancer cell proliferation, cycle, and apoptosis through regulating SOCS3/JAK-STAT signaling pathway. Immunopharmacol Immunotoxicol. 2018;40(4):278–283. doi:10.1080/08923973.2018.1455208
29. Gao X, Sun J, Huang C, Hu X, Jiang N, Lu C. RNAi-mediated silencing of NOX4 inhibited the invasion of gastric cancer cells through JAK2/STAT3 signaling. Am J Transl Res. 2017;9(10):4440.
30. Sethi G, Sung B, Aggarwal BB. Therapeutic potential of VEGI/TL1A in autoimmunity and cancer. In: Therapeutic Targets of the TNF Superfamily. Springer; 2009:207–215.
31. Zhang S, Yu D. Targeting Src family kinases in anti-cancer therapies: turning promise into triumph. Trends Pharmacol Sci. 2012;33(3):122–128. doi:10.1016/j.tips.2011.11.002
32. Elsberger B, Paravasthu D, Tovey S, Edwards J. Shorter disease-specific survival of ER-positive breast cancer patients with high cytoplasmic Src kinase expression after tamoxifen treatment. J Cancer Res Clin Oncol. 2012;138(2):327–332. doi:10.1007/s00432-011-1096-8
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.