Back to Journals » OncoTargets and Therapy » Volume 12
TMEFF2 inhibits pancreatic cancer cells proliferation, migration, and invasion by suppressing phosphorylation of the MAPK signaling pathway
Authors Han H, Zhan Z, Xu J, Song Z
Received 30 March 2019
Accepted for publication 30 May 2019
Published 23 December 2019 Volume 2019:12 Pages 11371—11382
DOI https://doi.org/10.2147/OTT.S210619
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Hongchao Han,*,1,2 Zhilin Zhan,*,3 Jie Xu,2 Zhenshun Song1
1Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Shanghai No. 10 People’s Hospital, Nanjing Medical University, Nanjing, People’s Republic of China; 2Department of General Surgery, Yancheng Third People’s Hospital, Yancheng, People’s Republic of China; 3Department of Hepatobiliary Surgery, Chizhou People’s Hospital, Chizhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhenshun Song
Department of Hepatobiliary and Pancreatic Surgery, The Affiliated Shanghai No. 10 People’s Hospital, Nanjing Medical University, No. 301 Yanchang Middle Road, Shanghai 200072, People’s Republic of China
Tel +86 137 6120 5962
Email [email protected]
Purpose: This paper studied the effect of TMEFF2 expression on pancreatic cancer and its mechanism.
Methods: A total of 72 pancreatic cancer patients were enrolled. AsPC1 and Panc1 cells were transfected. SB203580 was used to treat AsPC1 cells. CCK8 assay, colony formation analysis, Transwell experiment and Tunel test were performed. In vivo studies in nude mice were conducted. Immunohistochemistry, qRT-PCR and Western blot were used to detect genes expression.
Results: TMEFF2 was downregulated in pancreatic cancer tissues and cells (P<0.001). Low TMEFF2 expression was associated with larger tumor size and advanced stage and poor differentiation (P<0.01). Compared with the NC group, AsPC1 and Panc1 cells of the TMEFF2 group exhibited much lower OD450 values, colony number, tumor volume and weight, migration and invasion cell numbers, obviously higher E-cadherin protein expression, lower Snail, Vimentin, MMP-2 and MMP-9 proteins expression, lower phosphorylation level of MAPK signaling pathway, and more apoptotic cells. AsPC1 cells of the SB203580 group showed much lower OD450 value when compared with the siTMEFF2 group. Significantly decreased colony number, migration and invasion number, higher E-cadherin protein expression and lower Snail, Vimentin, MMP-2 and MMP-9 proteins expression were found in AsPC1 cells of the siTMEFF2+ SB203580 group when compared with the siTMEFF2+ DMSO group.
Conclusion: TMEFF2 inhibits pancreatic cancer cells proliferation, migration, and invasion by suppressing the phosphorylation of the MAPK signaling pathway.
Keywords: pancreatic cancer, TMEFF2, MAPK signaling, proliferation, migration
Introduction
Pancreatic cancer is a common malignant tumor of the digestive system, characterized by insidious onset, high degree of malignancy, rapid development and poor prognosis, with a five-year survival rate of less than 6%.1,2 In Western countries, pancreatic cancer is already the fourth leading cause of malignant tumor–related death, and by 2030, pancreatic cancer is expected to be the second leading cause of cancer-related death in the United States.3,4 Surgical resection combined with chemotherapy is still the main clinical treatment for pancreatic cancer.5 However, multiple complications and insensitivity to chemotherapy drugs have become the main causes of recurrence and metastasis.6,7 Therefore, a deep understanding of the molecular mechanisms underlying the occurrence and development of pancreatic cancer to find effective strategies for the treatment of pancreatic cancer is imminent.
TMEFF2, a novel member of the EGF-like protein family, was found to possess the characteristics of tumor suppressor in recent years because it inhibited proliferation of tumor cells in vitro and growth of tumors in nude mice.8,9 TMEFF2 is a transmembrane protein with EGF-like and two follistatin-like domains.10 Recent research indicated that TMEFF2 was obviously downregulated in gastric cancer patients, which was closely associated with poor prognosis such as large tumor size and advanced clinical stage.11 Green et al12 illustrated that TMEFF2 overexpression might inhibit sarcosine-induced invasion by interacting with sarcosine dehydrogenase. The tumor suppressive effect of TMEFF2 in prostate cancer has also been confirmed, and researchers explained that TMEFF2 might inhibit prostate cancer cells migration and invasion by modulating integrin expression and RhoA activation.13,14 Suarez et al15 reported low expression of TMEFF2 in glioma, which was associated with poor prognosis of glioma patients.
Overall, relatively few studies on TMEFF2 in human diseases have been discovered, and in the last 5 years, research about the effects of TMEFF2 expression on human tumors were also rarely emerged. Currently, no studies have been found on the effects of TMEFF2 expression on the progression of pancreatic cancer. This article firstly explored TMEFF2 expression in pancreatic cancer and its impact on the development of pancreatic cancer, aiming to provide a potential target for the targeted treatment of pancreatic cancer.
Methods
Patients and tissue specimens
A total of 72 patients who were diagnosed with pancreatic cancer in the Affiliated Shanghai No. 10th People’s Hospital, Nanjing Medical University, from June 2016 to September 2018 were enrolled, and tumor tissues as well as adjacent normal tissues from these patients were collected during surgery. The clinical information (including age, gender, tumor size, clinical stage, distant metastasis and differentiation) of these patients is listed in Table 1. This study has obtained written informed consent from all patients and has been approved by the ethics committee of the Affiliated Shanghai No. 10th People’s Hospital, Nanjing Medical University, and complied with the Declaration of Helsinki.
 |
Table 1 The relationship between TMEFF2 expression and clinical features |
Cell lines
Human normal pancreatic ductal epithelial cell line (HPDE6-C7) and pancreatic cancer cell lines (SW1990, Bxpc3, CFPAC1, Panc1 and AsPC1) were purchased from the cell bank of Peking Union Medical College (Beijing, China). DMEM containing 10% fetal bovine serum (FBS), 100 units/mL of penicillin and 100 μg/mL of streptomycin was prepared to culture cells in 25-mL sterile culture flasks at 37°C, 5% CO2, in an incubator. The DMEM in each flask was changed every two days. After three passages, cells were harvested in the logarithmic growth phase.
Transfection
AsPC1 and Panc1 cells in the logarithmic growth phase were separately prepared as single cell suspension (1 × 105 cells/mL) by using DMEM (without FBS). Each cell line was inoculated separately in 6-well plates with 1 mL cell suspension per well. After 24-hr incubation at 37°C, 5% CO2 in the incubator, cells in each well were transfected by pCDNA3.1-TMEFF2 overexpression vector (TMEFF2 group) and pCDNA3.1 empty vector (NC group), respectively. In addition, AsPC1 cells were also subjected to transfection with TMEFF2 siRNA (siTMEFF2 group) and negative control (siNC group). All plates were placed in the incubator for 6 hrs at 37°C, 5% CO2 conditions. The residual liquid in each well was discarded and replaced with fresh DMEM containing 10% FBS. All cells in each plate had undergone 48-hr culture at 37°C, 5% CO2 in the incubator.
Cell treatment with SB203580
AsPC1 cells of siNC group and siTMEFF2 group were seeded in 6-well plates with a density of 1 × 105 cells per well. Each well contained 1 mL DMEM (with 10% FBS). SB203580 (Calbiochem, San Diego, CA), dissolved in DMSO, was added to AsPC1 cells of siTMEFF2 group to a final concentration of 40 μM [16], and these cells were set as siTMEFF2+ SB203580 group. Equivalent volume of DMSO was also used to treat AsPC1 cells of siNC group and siTMEFF2 group along with being named siNC + DMSO group and siTMEFF2+ DMSO group. Meanwhile, AsPC1 cells without any treatment were also inoculated in 6-well plates with 1 × 105 cells and 1 mL DMEM (with 10% FBS) in each well. SB203580 dissolved in DMSO was then added into each well to a final SB203580 concentration of 40 μM (SB203580 group). All plates were placed in the incubator at 37°C, 5% CO2 surroundings.
CCK8 assay
Cells in logarithmic growth phase were harvested and seed in 96-well plates with 2 × 103 cells each well. A total of 100 μL DMEM containing 10% FBS was added to each well. Six duplicate wells were set in each group. All plates were placed in the sterile incubator for 0-, 24-, 48- and 72-hr incubation, respectively, at 37°C, 5% CO2 conditions. The culture medium in each well was changed once a day. At each time point, plates of each group were removed from the incubator and 10 μL of CCK8 solution (5 mg/mL) was added to each well. Plates were then returned to the incubator for 1-hr incubation. At 450-nm wavelength, the OD of each well was measured by a microplate reader (Biotek instrument, USA).
Colony formation analysis
A total of 5 mL single-cell suspensions of each group was inoculated into 600 culture dishes at a density of 1 × 103 cells/mL. All of these culture dishes were placed in the incubator for 2 weeks incubation at 37°C, 5% CO2. The culture medium in the culture dishes was changed every 3 days. Two weeks later, culture dishes were removed from the incubator and 4% paraformaldehyde was used to fix cells for 15 mins. Cells were then subjected to 0.1% crystal violet staining for 10 mins. All culture dishes were placed under a microscope and colony number of each group was counted with five non-overlapping fields. More than 50-cell aggregation was considered as one colony.
In vivo studies
Twelve nude mice (Shanghai Experimental Animal Center, Chinese Academy of Sciences) were kept in a sterile room at 25°C and were given free access to water and diet. In this study, all animal experiments have been approved by the Animal Ethics Committee of the Affiliated Shanghai No. 10th People’s Hospital, Nanjing Medical University,and were performed in accordance with the guidelines of the National Institutes of Health for the Care and Use of Laboratory Animals.
AsPC1 cell suspensions (dispersed in PBS, 1 × 105 cells/mL, 100 μL) of NC group and TMEFF2 group were subcutaneously injected into the back of nude mice. Each cell suspension was randomly injected with 6 nude mice. All nude mice were continued to be reared in the original environment. On the 7th, 14th, 21st, 28th and 35th day of the injection, the long (a) and short (b) diameter of subcutaneous tumors were measured using a vernier caliper to calculate tumor volume using the formula of (a × b2 × π)/6. On the 35th day, subcutaneous tumor tissues were collected after nude mice were sacrificed and tumor weight was detected.
Transwell experiment
The upper chamber of the Transwell chamber with a pore size of 8 μm was coated with a layer of 100 μL Matrigel to detect the invasion ability of cells. It should be noted that, in the detection of cells migration ability, Matrigel was not be allowed to be placed in the upper chamber of the Transwell chamber. Cells in the logarithmic growth phase were prepared as single-cell suspensions using DMEM serum-free medium (1 × 105 cells/mL). A total of 100 μL cell suspensions were added to the upper layer of the chamber. The lower chamber contained 600 μL DMEM medium containing 10% FBS. All Transwell chambers were placed in the incubator for 24 hrs at 37°C, 5% CO2. Then, these Transwell chambers were removed from the incubator, and cells remaining on the upper membrane of the chamber were gently scraped off using a cotton swab. Formaldehyde (4%) was used to fix cells that passed through the membrane. These cells then underwent crystal violet staining for 10 mins and were observed under a microscope. Five non-overlapping fields were randomly selected to count the number of cells passing through the membrane.
Immunohistochemistry
Tumor tissues were fixed in formalin, embedded in paraffin and cut into slices of 4-μm thickness. All tumor sections were subjected to dewaxing and hydration using xylene and gradient ethanol. After being subjected to antigen retrieval using 0.01 M citrate buffer, sections were washed with PBS for 3 times and then incubated with 3% H2O2 for 10 mins to remove endogenous peroxidase activity. Washing 3 times with PBS was still necessary for these tissues. Goat serum was used to block sections for 30 mins. Mouse anti-human Ki67 monoclonal antibody (1:100, Fuzhou Maixin Biotechnology Development Co. Ltd, Fuzhou, China) and mouse anti-human TMEFF2 monoclonal antibody (1:100, Novus Biologicals, Littleton, Colorado, USA) were added to the sections. All sections were placed in a refrigerator for overnight incubation at 4°C. After washing 3 times with PBS, the sections were incubated with horseradish peroxidase–labeled secondary antibody (1:500, Biogenex, San Ramon, CA, USA) for 30 mins at room temperature. PBS was used to rinse sections for 3 times for 5 mins each rinse. DAB color reaction and hematoxylin counterstaining were performed on the sections. Subsequently, all the sections were dehydrated and sealed with neutral resin. TMEFF2 or Ki67-positive expression cells were observed and counted under a microscope with 5 random non-overlapping fields. Cells with brownish yellow particles were considered positive expression cells.
Tunel test
Dewaxed tumor tissue sections were incubated with 0.2% Triton X-100 for 10 mins and then incubated with 45 μL Tunel test solution for 1 hr at room temperature in the dark. After washing 3 times with PBS, the sections were observed under a fluorescence microscope. Five tumor sections were randomly selected from each tumor tissue and 3 non-overlapping fields of each section were randomly selected to count the number of Tunel-positive cells.
qRT-PCR
This research detected the TMEFF2 mRNA expression by using qRT-PCR. Biefly, total RNA in tissues and cells was collected using RNeasy mini kit (Qiagen, Valencia, CA). A total of 1 μg RNA sample was subjected to reverse transcription reaction using SuperScript III First-Strand synthesis system (Life Technologies, Grand Island, NY) in order to obtain cDNA template. The PCR program was set as follows: 95°C for 5 mins and 40 cycles of 95°C for 1 min, 58°C for 1 min and 72°C for 30 s. TaqMan Gene Expression Assay (Applied Biosystems, Carlsbad, CA) on the ABI 7300 Real-Time PCR system was used to carry out the abovementioned PCR program. Primer sequences were listed as follows: TMEFF2, forward, 5ʹ -CTGATGGGAAATCTTATGATAATG-3ʹ, reverse, 5ʹ -CAGGAACAACGTAGAGAACACTGT-3ʹ. β-actin (internal control), forward, 5ʹ-TTCGAGCAGGAATCTGACACAT-3ʹ, reverse, 5ʹ-CAATGATGGCTGGAAGAGGAC-3ʹ. Data was processed by 2−ΔΔCt method.
Western blot
Cells and tissues that were ground into powder in liquid nitrogen were lysed in ice-cold RIPA buffer (Beyotime, China). Through centrifugation at 4°C, impurities such as insoluble materials and cell debris were removed. Protein samples were obtained and boiled with loading buffer at 100°C. Totally, 80 µg of each protein sample was subjected to 2 hrs of SDS-PAGE electrophoresis, followed by being transferred to a PVDF membrane at a voltage of 15 V. The PVDF membrane was incubated for 1 hr at room temperature in 5% skim milk powder and was placed in the refrigerator overnight at 4°C to incubate with primary antibodies (mouse anti-TMEFF2, 1:1000, R&D Systems, Wiesbaden, Germany; mouse anti-E-cadherin, 1:1000, BD Biosciences, Palo Alto, CA; rabbit anti-Snail, 1:1000, Imgenex, San Diego, CA, USA; mouse anti-Vimentin, 1:1000, Cell Signaling Technology, Danvers, MA, USA; rabbit anti-MMP-2 and rabbit anti-MMP-9, 1:1000, Bioworld Tech., MN, USA; rabbit anti-p-JNK, rabbit anti-JNK, rabbit anti-p-P38 and rabbit anti-P38, 1:1000, Cell Signaling Technology, Danvers, MA, USA; mouse anti-β-actin, 1:1000, Sigma, St. Louis, MO, USA; rabbit anti-Ras, rabbit anti-Raf, rabbit anti-p-Raf, rabbit anti-MEK, rabbit anti-p-MEK, rabbit anti-ERK and rabbit anti-p-ERK, 1:1000, BD Biosciences, Palo Alto, CA). TBST was used to wash the membrane, and horseradish peroxidase-conjugated secondary antibody (1:5000, Dako, Denmark) was selected to incubate the membrane for 2 hrs at room temperature. At last, immune blot signals were detected by ECL system (Thermo Fisher, USA). β-actin was served as an internal control.
Statistical analysis
In this paper, all experiments were performed independently 3 times. Data was exhibited in a form of mean ± SD with SPSS 19.0 (SPSS Inc., Chicago, IL, USA) as the processing software. Student’s t-test or one-way ANOVA was used to analyze data from two or multiple groups, respectively. Pearson’s χ2 test was used to analyze the correlation between clinicopathological features and TMEFF2 expression in pancreatic cancer patients. P<0.05 meant statistically significant.
Results
Low expression of TMEFF2 in pancreatic cancer was associated with poor outcome of patients
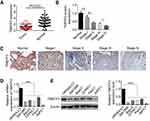
TMEFF2 mRNA expression in tumor tissues and adjacent normal tissues of 72 pancreatic cancer patients were detected, and significantly lower TMEFF2 mRNA expression was found in tumor tissues when compared with that in normal tissues (P<0.001) (Figure 1A). We also analyzed the relationship between TMEFF2 protein expression level and patients’ clinical features. As shown in Table 1, it could be noted that gender, age and distant metastasis did not significantly affect the expression of TMEFF2 protein. However, tumor size, clinical stage and differentiation had obviously effect on TMEFF2 protein expression level. For patients with larger tumor size, advanced stage and poor differentiation, the TMEFF2 protein expression in their tumor tissues was much higher than those with smaller tumor size, early stage, and moderate and well differentiation (P<0.05 or P<0.01).
We have a more detailed distinction on clinical stages of patients. Compared with normal tissues, much reduced TMEFF2 protein expression was found in tumor tissues of patients with stage I (P<0.01). Tumor tissues of patients with stage II showed markedly lower TMEFF2 protein expression than those with stage I (P<0.01). No significant difference was found in TMEFF2 protein expression between patients with stage II and stage III. However, obviously decreased TMEFF2 protein expression was observed in tumor tissues of patients with stage IV when compared with patients with stage III (P<0.05) (Figure 1B). Immunohistochemistry results also revealed that TMEFF2-positive cell numbers in tumor tissues were gradually decreased with the increase of clinical stage (Figure 1C).
Through in vitro studies, we further verified the expression of TMEFF2 in pancreatic cancer cells and found that the relative TMEFF2 mRNA and protein expressions were much declined in pancreatic cancer cells (SW1990, Bxpc3, CFPAC1, Panc1 and AsPC1) than that in human normal pancreatic ductal epithelial cells (HPDE6-C7) (P<0.001) (Figure 1D and E). AsPC1 cells with a lower TMEFF2 expression and Panc1 cells with a higher TMEFF2 expression were selected for follow-up studies.
TMEFF2 inhibited pancreatic cancer cells in vitro proliferation and in vivo growth
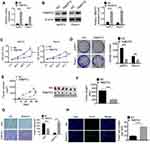
After transfection, the relative TMEFF2 mRNA and protein expressions in AsPC1 and Panc1 cells of TMEFF2 group were significantly decreased than that of NC group, indicating that cells were successfully transfected (P<0.001) (Figure 2A and B). According to CCK8 assay, after 72 hrs of transfection, the OD450 values of AsPC1 and Panc1 cells in TMEFF2 group were dramatically lower than that in NC group (P<0.001) (Figure 2C). Results from colony formation analysis showed much lower colony number of TMEFF2 group when compared with NC group (P<0.01 or P<0.001) (Figure 2D).
In vivo experiments in nude mice were conducted by subcutaneous injection of pancreatic cancer single-cell suspensions. After subcutaneous injection for 35 d, remarkably lower tumor volume and tumor weight occurred in TMEFF2 group when compared with NC group (P<0.001) (Figure 2E and F). Immunohistochemistry of subcutaneous tumor tissues exhibited that tumor tissues of TMEFF2 group showed much higher positive TMEFF2 expression cell number and obviously lower negative Ki67 expression cell number than that of NC group (Figure 2G). In addition, the apoptosis of tumor tissue cells was also detected by Tunel test, and significantly higher Tunel+ cell number was found in tumor tissues of TMEFF2 group when compared with that of NC group (P<0.001) (Figure 2H).
TMEFF2 inhibited pancreatic cancer cell migration and invasion and epithelial-mesenchymal transition (EMT)
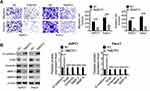
Migration and invasion abilities of AsPC1 and Panc1 cells in NC group and TMEFF2 group were explored by Transwell experiment. Compared with the number of migration and invasion cells in NC group, it was significantly decreased in TMEFF2 group (P<0.001) (Figure 3A). EMT-related protein expression in cells of the two groups was also measured by Western blot. As shown in Figure 3B, compared with NC group, the relative E-cadherin protein expression in AsPC1 and Panc1 cells of TMEFF2 group was dramatically increased (P<0.001). However, remarkably decreased relative Snail, Vimentin, MMP-2 and MMP-9 protein expression was observed in AsPC1 and Panc1 cells of TMEFF2 group when compared with NC group (P<0.001).
TMEFF2 inhibited pancreatic cancer cell proliferation, migration, invasion and EMT by suppressing the phosphorylation of MAPK signaling pathway
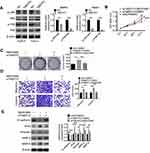
JNK and P38 were two important proteins in the MAPK signaling pathway, and the activation of MAPK signaling pathway had important impacts on tumor development. In this research, the expression of JNK, p-JNK, P38 and p-P38 proteins was detected in AsPC1 and Panc1 cells of NC group and TMEFF2 group, and the ratio of p-JNK/JNK and p-P38/P38 was calculated according to the measured expression levels of each protein. As a result, AsPC1 and Panc1 cells of TMEFF2 group had a much lower ratio of p-JNK/JNK and p-P38/P38 when compared with NC group (P<0.01 or P<0.001) (Figure 4A). SB203580, an inhibitor of the MAPK signaling pathway, 16 was used to treat AsPC1 cells to investigate whether TMEFF2 affected pancreatic cancer cells' biological behavior by affecting the MAPK signaling pathway. As shown in Figure 4B, compared with siNC + DMSO group, AsPC1 cells of siTMEFF2+ DMSO group exhibited much higher OD450 value at 72 hrs (P<0.05). However, significantly decreased OD450 value was found in AsPC1 cells of siTMEFF2+ SB203580 group when compared with siTMEFF2 group (P<0.001).
Results from colony formation analysis and Transwell experiment exhibited that compared with siNC + DMSO group, AsPC1 cells of siTMEFF2+ DMSO group had obviously higher colony number and migration and invasion cell numbers (P<0.01 or P<0.001). However, significantly reduced colony number and migration and invasion cell numbers were found in AsPC1 cells of siTMEFF2+ SB203580 group when compared with siTMEFF2+ DMSO group (P<0.001) (Figure 4C and D). Furthermore, much lower E-cadherin protein expression and obviously higher Snail, Vimentin, MMP-2 and MMP-9 proteins expression were observed in AsPC1 cells of siTMEFF2+ DMSO group when compared with that of siNC + DMSO group (P<0.01 or P<0.001). However, when compared with siTMEFF2+ DMSO group, AsPC1 cells of siTMEFF2+ SB203580 group had significantly higher E-cadherin protein expression and much lower Snail, Vimentin, MMP-2 and MMP-9 proteins expression (P<0.05 or P<0.01 or P<0.001) (Figure 4E).
TMEFF2 inhibited the phosphorylation of Ras/Raf/MEK/ERK signaling pathway in pancreatic cancer cells
The effects of TMEFF2 expression on the Ras/Raf/MEK/ERK signaling pathway were researched and results are shown in Figure 5. It could be discovered that compared with NC group, prominently reduced expression of Ras, p-Raf/Raf, p-MEK/MEK, p-ERK/ERK occurred in AsPC1 and Panc1 cells of TMEFF2 group (P<0.001). It was well known that Ras/Raf/MEK/ERK signaling pathway was involved in the development of a variety of tumors, including pancreatic cancer. Thus, it could be concluded that TMEFF2 inhibited the progression of pancreatic cancer by interfering with the phosphorylation of Ras/Raf/MEK/ERK signaling pathway.
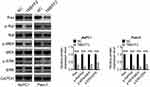 |
Figure 5 TMEFF2 inhibited the phosphorylation of Ras/Raf/MEK/ERK signaling pathway in pancreatic cancer cells. ***P<0.001. Abbreviation: NC, negative control. |
Discussion
In the past few decades, molecular-targeted therapy had become a hot spot in clinical oncology research, and the discovery of some effective potential therapeutic targets was expected to completely cure human tumors in the future. Pancreatic cancer is a common malignant tumor that causes a serious poor prognosis for patients, and some potential molecular therapeutic targets for pancreatic cancer have also emerged.17–20 In this study, TMEFF2 protein expression in tumor tissues of pancreatic cancer patients was detected using Western blot and immunohistochemistry, and downregulated TMEFF2 protein expression was found to be associated with poor prognosis. Overexpression of TMEFF2 could inhibit pancreatic cancer cell proliferation in vitro and growth in vivo. Some existing analyses of TMEFF2 in human tumors suggested that TMEFF2 was a tumor suppressor gene and it could suppress prostate cancer cells growth.21 Sun et al22 indicated that TMEFF2 expression was significantly decreased in gastric cancer cells induced by Helicobacter pylori, and overexpression of TMEFF2 might contribute to Helicobacter pylori–induced gastric carcinogenesis by negatively regulating phosphorylation of STAT3. Our data also revealed that TMEFF2 was a tumor suppressor in pancreatic cancer. To the best of our knowledge, this is the first time TMEFF2 has been studied in pancreatic cancer. In addition, according to immunohistochemistry, TMEFF2 overexpression inhibited the expression level of Ki67 in transplanted tumor tissues of nude mice. Ki67 is a widely recognized cell proliferation marker that regulates the cell cycle by affecting cell mitosis, thereby promoting tumor cell proliferation and growth.23–25
In the EMT process, epithelial-derived malignant cells will be transformed into more migrating and invasive interstitial cells, which greatly promote the malignant development of tumors.26 Decreased adhesion between cells, rupture of cell desmosome and hemidesmosome, loss of cell polarity, secretion of matrix metalloproteinase and degradation of extracellular matrix are the main features of EMT, which enable tumor cells to gain greater exercise capacity and more space for migration and invasion.27 E-cadherin, Snail, Vimentin, MMP-2 and MMP-9 proteins are all EMT-related proteins, which play an irreplaceable role in the occurrence of EMT.28 The main marker of EMT is the decrease in E-cadherin expression, and Snail binds to the E-box element on the E-cadherin promoter to inhibit the transcription of E-cadherin, thereby resulting in loss of adhesion between tumor cells and enhanced ability to invade and metastasize.29 Vimentin is a major component of intermediate fibers, and its high expression has been confirmed to enhance the invasive ability of tumor cells.30 MMP-2 and MMP-9, members of the matrix metalloproteinase family, can destroy the basement membrane by degrading IV, V-type collagen and various matrix components, thereby causing tumor cells to infiltrate and metastasize to other parts of the human body.31 Data from this paper indicated that overexpression of TMEFF2 attenuated pancreatic cancer cells migration and invasion by inhibiting EMT. After TMEFF2 was overexpressed, upregulated E-cadherin protein expression and downregulated Snail, Vimentin, MMP-2 and MMP-9 protein expression were found in pancreatic cancer cells.
MAPK signaling pathway has been reported to be involved in the development of various human tumors. In this research, we noticed that TMEFF2 upregulation could significantly inhibit the phosphorylation of JNK and P38 in pancreatic cancer cells. JNK and P38 are two important components of the MAPK signaling pathway, and their phosphorylation greatly promotes the progression of tumors.32 Previous studies have also demonstrated that the phosphorylation of JNK and P38 promoted pancreatic cancer cell proliferation, migration and invasion.33,34 Existing literature documented that TMEFF2 could participate in the regulation of human diseases development by regulating important signaling pathways, such as AKT, ERK and CRH signaling pathways.9,35 This is the first time that TMEFF2 has been found to inhibit the proliferation, migration and invasion of pancreatic cancer cells by suppressing the phosphorylation of MAPK signaling pathway. In addition, we also discovered that TMEFF2 overexpression inhibited the phosphorylation of Ras/Raf/MEK/ERK signaling pathway in pancreatic cancer cells. The Ras/Raf/MEK/ERK signaling pathway was one of the most classical pathway in the MAPK pathways, which was widely involved in the regulation of cell biological behavior and was closely related to the malignant phenotype of tumor cells.36 After the Ras/Raf/MEK/ERK signaling pathway was activated, the expression of Ras, p-Raf, p-MEK and p-ERK protein was increased, resulting in increased cyclin expression and elevated levels of CDK-Cyclin complex. Cells were then induced from the G0/G1 phase to the S phase, thereby promoting the progression of the cell cycle.37 Previous studies also illustrated that the inhibition of Ras/Raf/MEK/ERK signaling pathway activity could impede the progression of pancreatic cancer.38,39 Our data also indicated that TMEFF2 might inhibit the progression of pancreatic cancer by interfering with the phosphorylation of Ras/Raf/MEK/ERK signaling pathway.
There were limitations in this study. A more complete genotype and phenotype for each cell lines should be researched and whether TMEFF2 expression depending on TP53 mutation status should also be explored. However, due to limitations of our laboratory conditions, we are currently unable to conduct in-depth researches on this point and this will be the focus of our future research.
In short, this study discovered that TMEFF2 was significantly downregulated in pancreatic cancer and low TMEFF2 expression was obviously associated with poor outcome of patients with pancreatic cancer. More importantly, overexpression of TMEFF2 might inhibit pancreatic cancer cell proliferation, migration and invasion by suppressing the phosphorylation of MAPK signaling pathway. With the emergence of more relevant research in the future, TMEFF2 is expected to become an effective target for the treatment of pancreatic cancer.
Highlights
- Low TMEFF2 expression in pancreatic cancer was associated with poor outcome.
- TMEFF2 inhibited pancreatic cancer cells in vitro proliferation and in vivo growth.
- TMEFF2 inhibited pancreatic cancer cells migration and invasion.
- TMEFF2 inhibited MAPK signaling pathway activity in pancreatic cancer cells.
Disclosure
The authors report no conflicts of interest in regard to this work.
References
1. Beeghly-Fadiel A, Luu HN, Du L, et al. Early onset pancreatic malignancies: clinical characteristics and survival associations. Int J Cancer. 2016;139(10):2169–2177. doi:10.1002/ijc.30273
2. Camara, Soriba SN, Yin T, et al. High risk factors of pancreatic carcinoma. J Huazhong Univ Sci Technolog Med Sci. 2016;36(3):295–304. doi:10.1007/s11596-016-1583-x
3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65(1):5–29. doi:10.3322/caac.21254
4. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
5. Saung MT, Zheng L. Current standards of chemotherapy for pancreatic cancer. Clin Ther. 2017;39(11):S0149291817309049. doi:10.1016/j.clinthera.2017.08.015
6. Li D, Mullinax JE, Aiken T, et al. Loss of PDPK1 abrogates resistance to gemcitabine in label-retaining pancreatic cancer cells. BMC Cancer. 2018;18(1):772. doi:10.1186/s12885-018-4242-8
7. Watanabe Y, Nishihara K, Matsumoto S, Okayama T, Abe Y, Nakano T. Effect of postoperative major complications on prognosis after pancreatectomy for pancreatic cancer: a retrospective review. Surg Today. 2016;47(5):1–13. doi:10.1007/s00595-016-1321-9
8. Chen X, Overcash R, Green T, Hoffman D, Asch AS, Ruiz-Echevarría MJ. The tumor suppressor activity of the transmembrane protein with epidermal growth factor and two follistatin motifs 2 (TMEFF2) correlates with its ability to modulate sarcosine levels. J Biol Chem. 2011;286(18):16091–16100. doi:10.1074/jbc.M110.193805
9. Labeur M, Woelfel B, Stalla J, Gk S. TMEFF2 is an endogenous inhibitor of the CRH signal transduction pathway. J Mol Endocrinol. 2015;54(1):51. doi:10.1530/JME-14-0225
10. Huang H, Teng P, Mei R, et al. Tmeff2 is expressed in differentiating oligodendrocytes but dispensable for their differentiation in vivo. Sci Rep. 2017;7(1):337. doi:10.1038/srep41904
11. Tiantian S, Du W, Chen HY, Hong J, Fang JY. TMEFF2 deregulation contributes to gastric carcinogenesis and indicates poor survival outcome. Clin Gastroenterol Hepatol. 2015;13(7):e79–e79. doi:10.1016/j.cgh.2015.04.055
12. Green T, Chen X, Ryan S, Asch AS, Ruiz-Echevarría MJ. TMEFF2 and SARDH cooperate to modulate one-carbon metabolism and invasion of prostate cancer cells. Prostate. 2013;73(14):1561–1575. doi:10.1002/pros.22706
13. Corbin JM, Overcash RF, Wren JD, et al. Analysis of TMEFF2 allografts and transgenic mouse models reveals roles in prostate regeneration and cancer. Prostate. 2016;76(1):97–113. doi:10.1002/pros.23103
14. Chen X, Corbin JM, Tipton GJ, Yang LV, Asch AS, Ruiz-Echevarría MJ. The TMEFF2 tumor suppressor modulates integrin expression, RhoA activation and migration of prostate cancer cells ☆. BBA Mol Cell Res. 2014;1843(6):1216–1224. doi:10.1016/j.bbamcr.2014.03.005
15. Suarez EG, Carneiro C, Torrecilla D, et al. 859 TGF-b inhibits TMEFF2 expression in glioma cells. Eur J Cancer. 2012;48(Suppl. 5):S207–S207. doi:10.1016/S0959-8049(12)71492-2
16. Lin Z, Crockett DK, Jenson SD, Lim MS, Elenitoba-Johnson KSJ. Quantitative proteomic and transcriptional analysis of the response to the p38 mitogen-activated protein kinase inhibitor SB203580 in transformed follicular lymphoma cells. Mol Cell Proteomics. 2004;3(8):820. doi:10.1074/mcp.M400008-MCP200
17. Guo S, Xu X, Ouyang Y, et al. Microarray expression profile analysis of circular RNAs in pancreatic cancer. Mol Med Rep. 2018;17(6):7661–7671. doi:10.3892/mmr.2018.8827
18. Qin CF, Zhao FL. Long non-coding RNA TUG1 can promote proliferation and migration of pancreatic cancer via EMT pathway. Eur Rev Med Pharmacol Sci. 2017;21(10):2377.
19. Okura R, Fujihara S, Iwama H, et al. MicroRNA profiles during galectin-9-induced apoptosis of pancreatic cancer cells. Oncol Lett. 2018;15(1):407–414. doi:10.3892/ol.2017.7316
20. Wang L, Shureiqi I, Stroehlein JR, Wei D. Novel and emerging innate immune therapeutic targets for pancreatic cancer. Expert Opin Ther Targets. 2018;22:12. doi:10.1080/14728222.2018.1538361
21. Gery S, Sawyers CL, Agus DB, Said JW, Koeffler HP. TMEFF2 is an androgen-regulated gene exhibiting antiproliferative effects in prostate cancer cells. Oncogene. 2002;21(31):4739–4746. doi:10.1038/sj.onc.1205142
22. Tian-Tian S, Tang J-Y, Du W, et al. Bidirectional regulation between TMEFF2 and STAT3 may contribute to Helicobacter pylori-associated gastric carcinogenesis. Int J Cancer. 2015;136(5):1053–1064. doi:10.1002/ijc.29061
23. Thakur SS, Li H, Chan AMY, et al. The use of automated Ki67 analysis to predict Oncotype DX risk-of-recurrence categories in early-stage breast cancer. PLoS One. 2018;13(1):e0188983. doi:10.1371/journal.pone.0188983
24. Li LT, Jiang G, Chen Q, Zheng JN. Ki67 is a promising molecular target in the diagnosis of cancer (review). Mol Med Rep. 2015;11(3):1566–1572. doi:10.3892/mmr.2014.2914
25. Yuan JP, Wang LW, Qu AP, et al. Quantum dots-based quantitative and in situ multiple imaging on Ki67 and cytokeratin to improve Ki67 assessment in breast cancer. PLoS One. 2015;10(4):e0122734. doi:10.1371/journal.pone.0122734
26. Burute M, Prioux M, Blin G, et al. Polarity reversal by centrosome repositioning primes cell scattering during epithelial-to-mesenchymal transition. Dev Cell. 2017;40(2):168–184. doi:10.1016/j.devcel.2016.12.004
27. Hervé A, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions: the importance of changing cell state in development and disease. Cell. 2009;139(5):871. doi:10.1016/j.cell.2009.11.007
28. Tuo Z, Zhang J, Xue W. LncRNA TP73-AS1 predicts the prognosis of bladder cancer patients and functions as a suppressor for bladder cancer by EMT pathway. Biochem Biophys Res Commun. 2018;499:875–881. doi:10.1016/j.bbrc.2018.04.010
29. Papiewska-Pająk I, Kowalska MA, Boncela J. Expression and activity of SNAIL transcription factor during epithelial to mesenchymal transition (EMT) in cancer progression. Postepy Hig Med Dosw. 2016;70:968. doi:10.5604/17322693.1219401
30. Xu, Z., Bian H, Zhang F, et al. URI promotes the migration and invasion of human cervical cancer cells potentially via upregulation of vimentin expression. Am J Transl Res. 2017;9(6):3037.
31. Roomi MW, Kalinovsky T, Rath M, Niedzwiecki A. Modulation of MMP-2 and MMP-9 secretion by cytokines, inducers and inhibitors in human glioblastoma T-98G cells. Oncol Rep. 2017;37(3):1907–1913
32. Zhao M, Howard EW, Parris AB, Guo Z, Zhao Q, Yang X. Alcohol promotes migration and invasion of triple‐negative breast cancer cells through activation of p38 MAPK and JNK. Mol Carcinog. 2016;56(3):849. doi:10.1002/mc.v56.3
33. Liu H, Xu XF, Zhao Y, Tang MC, Zhou YQ, Gao FH. NS‑398 promotes pancreatic cancer cell invasion by CD147 and MMP‑2 via the activation of P38. Mol Med Rep. 2016;13:3.
34. Zhang X, Cao J, Pei Y, Zhang J, Wang Q. Smad4 inhibits cell migration via suppression of JNK activity in human pancreatic carcinoma PANC-1 cells. Oncol Lett. 2016;11(5):3465–3470. doi:10.3892/ol.2016.4427
35. Chen X, Ruizechevarría MJ. TMEFF2 modulates the AKT and ERK signaling pathways. Int J Biochem Mol Biol. 2013;4(2):83–94.
36. Asati V, Mahapatra DK, Bharti SK. PI3K/Akt/mTOR and Ras/Raf/MEK/ERK signaling pathways inhibitors as anticancer agents: structural and pharmacological perspectives. Eur J Med Chem. 2016;109:314–341. doi:10.1016/j.ejmech.2016.01.012
37. Yang S, Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma. Oncol Lett. 2017;13(3):1041. doi:10.3892/ol.2017.5557
38. Ulivi P, Arienti C, Amadori D, et al. Role of RAF/MEK/ERK pathway, p-STAT-3 and Mcl-1 in sorafenib activity in human pancreatic cancer cell lines. J Cell Physiol. 2010;220(1):214–221. doi:10.1002/jcp.v220:1
39. Zhang T, Li Y, Zhu Z, Gu M, Newman B, Sun D. MEK inhibition potentiates the activity of Hsp90 inhibitor 17-AAG against pancreatic cancer cells. Mol Pharm. 2010;7(5):1576–1584. doi:10.1021/mp900321a
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.