Back to Journals » OncoTargets and Therapy » Volume 12
TIMP1 Is A Potential Key Gene Associated With The Pathogenesis And Prognosis Of Ulcerative Colitis-Associated Colorectal Cancer
Authors Huang R, Wang K , Gao L, Gao W
Received 9 July 2019
Accepted for publication 9 October 2019
Published 30 October 2019 Volume 2019:12 Pages 8895—8904
DOI https://doi.org/10.2147/OTT.S222608
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Ru Huang,1,* Kaijing Wang,2,* Lei Gao,1 Wei Gao2
1Department of Heart Failure, Research Center for Translational Medicine, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China; 2Department of Colorectal Surgery, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Wei Gao
Department of Colorectal Surgery, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
Email [email protected]
Lei Gao
Department of Heart Failure, Research Center for Translational Medicine, Shanghai East Hospital, Tongji University School of Medicine, Shanghai, People’s Republic of China
Email [email protected]
Purpose: Colorectal cancer (CRC) is the third most frequently diagnosed cancer worldwide. As a high-risk factor for CRC, ulcerative colitis (UC) has been demonstrated to lead to epithelial dysplasia, DNA damage, and eventually cancer. There are approximately 18% of patients with UC may develop CRC.
Patients and methods: The gene expression profiles were retrieved from the Gene Expression Omnibus. The Database for Annotation, Visualization and Integrated Discovery was employed to conduct gene annotations. Protein-protein interaction network was constructed by the Search Tool for the Retrieval of Interacting Genes, and further analysed by the Molecular Complex Detection. The correlation between TIMP1 and prognosis was evaluated by the Gene Expression Profiling Interactive Analysis. To predict the potential functions of TIMP1, the GeneMANIA, Coremine, and FunRich were employed. After transfection with small interfering RNA targeting TIMP1, cell proliferation, migration, and apoptosis were determined by CCK-8, scratch wound, and Annexin V-FITC/PI assays, respectively.
Results: TIMP1, consistently overexpressed in the initiation and progression of UC-associated CRC (ucaCRC), was identified to be a potential biomarker for the prognosis of patients with CRC. Experimental results showed knockdown of TIMP1 could increase the migration, while did not affect the proliferation and apoptosis of RKO cells. The role of TIMP1 in the malignant transformation of ucaCRC was confirmed by using the protein/gene interactions and biological process annotation and validated by analysing the transcription factors targeting TIMP1.
Conclusion: TIMP1 is consistently upregulated in the pathological process of ucaCRC and can be a potential biomarker for the worse prognosis of CRC.
Keywords: colitis ulcerative, colorectal neoplasms, high-throughput screening assays, prognosis
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed cancer and also the second most common cancer-related death worldwide, which accounts for roughly 1.8 million new cases and 881,000 deaths in 2018.1 The therapeutic approaches, such as surgery and adjuvant therapy, have increased the overall survival rate for patients with CRC, however, the outcomes of these patients remain poor due to metastasis.2 There are several mechanisms leading to CRC such as genomic instability and mutations of tumor-suppressor genes,3 and also several high-risk factors for CRC such as hereditary nonpolyposis colon cancer syndrome, familial adenomatous polyposis, and ulcerative colitis (UC).4 UC is characterized by leukocyte infiltration and uncontrolled inflammatory reactions,5 and such pathologic features can lead to epithelial dysplasia, DNA damage, and eventually malignancy.6–8 Particularly, there are approximately 18% of the patients with UC may eventually develop CRC after 30 years of diseases.9 However, the molecular mechanisms driving the progression of ucaCRC have not been fully characterized yet.
To identify the potential genes that promote the progression of normal mucosa to ucaCRC, the microarray and high throughput sequencing technologies have been adopted recently.10,11 To overcome the limited or inconsistent results result from the different platforms or a small sample size, the integrated bioinformatics methods has been uncovered.12 As a result, through combining the datasets retrieved from the Gene Expression Omnibus (GEO) and subsequent bioinformatics analysis, we have previously identified several crucial genes functioning pivotally in many pathological processes including five candidates to be the potential diagnostic biomarkers, NMU-mediated alectinib resistance in non-small cell lung cancer, and COL4A1-mediated trastuzumab resistance in gastric cancer.13–15
Therefore, in the present study, through retrieving the datasets from the GEO and conducting an integrative bioinformatics analysis, we identify that TIMP metallopeptidase inhibitor 1, also known as TIMP1, upregulated consistently in the initiation and progression of ucaCRC may function pivotally in the malignant transformation of ucaCRC via multiple mechanisms.
Materials And Methods
Processing Of Microarray Data
In the present study, the gene expression profiles of GSE9452, GSE8671, GSE14538, GSE46451, and GSE31106 were retrieved from the GEO (http://www.ncbi.nlm.nih.gov/geo) as previously reported.15 In detail, GSE9452 contains 5 control samples and 21 UC samples;16 GSE8671 is consisted of 32 colorectal adenomas and the corresponding normal mucosa from the same individuals;17 GSE14538 contains one sample (Caco2 cell line) treated with 20 mmol/L mesalazine for 4 days and 1 untreated sample;18 GSE46451 is composed of 12 mucosal biopsies including 3 biological replicates in each of 4 categories (non-inflamed with or without mesalazine, inflamed with or without mesalazine);19 GSE31106 includes 3 colorectal tissues collected from AOM/DSS-induced CRC mice model at inflamed stage, dysplasia stages (low-grade dysplasia, high grade dysplasia and high-grade dysplasia with active inflammation) and carcinoma stage separately, and 3 normal colorectal tissues from control group.20 The detailed information of these datasets were provided in the supplementary materials. These microarray datasets were then analyzed by using the GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r/), an online tool used to compare two or more groups.21 P value < 0.05 and |log2FC| ≥ 1 were statistically considered as the cut-off criterion.
Functional And Pathway Enrichment Analyses
To obtain the functional annotation of these differentially expressed genes (DEGs), the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) was next employed to perform the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses.22 P < 0.05 was regarded as the significant threshold.
Construction Of The Protein-Protein Interaction Network (PPI)
As a widely used online database, the Search Tool for the Retrieval of Interacting Genes (STRING, http://string-db.org), has been applied to provide information regarding the predicted and experimental protein interaction.23 To explore the interactive associations between 78 significantly upregulated DEGs and 7 target genes of mesalazine, we mapped these genes to the STRING, and confidence score > 0.4 was set as significant. The Molecular Complex Detection (MCODE) was used to screen the modules of PPI network in the Cytoscape, and genes within the highest MCODE scores module are considered as hub genes.
Survival Analysis Of TIMP1
To evaluate the correlation between the expression of TIMP1 and its clinical outcomes for CRC, Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancerpku.cn/index.html), an online database based on The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) projects, was employed. Based on the expression level of TIMP1, CRC patients were divided into low and high expression groups respectively. The overall survival was then analyzed. The hazard ratio (HR) with 95% confidence intervals and log rank P value were computed.
GeneMANIA And Coremine Analyses
The GeneMANIA (https://www.genemania.org/) was applied to predict the potential functions of TIMP1 as introduced previously.24 Moreover, the Coremine Medical online database (https://www.coremine.com/medical/) was employed to conduct the annotations of biological processes involving TIMP1, CRC, and UC.
Analysis Of The Transcription Factors Targeting TIMP1
To look for the potential transcription factors targeting TIMP1, the FunRich (https://www.funrich.org), which is an analysis tool to identify the transcription factors for the specific genes, was also employed.25
Cell Culture And Transfection
RKO cells were obtained from the American Type Culture Collection (ATCC), and grown in Dulbecco’s modified Eagle’s medium (Gibco, USA) supplemented with 10% fetal bovine serum (Hyclone, UK), 100 U/mL penicillin and 100 µg/mL streptomycin (Gibco, USA). Cells were maintained at 37 °C in a 5% CO2 incubator. The siRNA against TIMP1 and the negative control oligo were designed following LNA Oligo Tools and Design Guidelines (Exiqon, Denmark), and synthesized by GenScript (Nanjing, China). The HiPerFect transfection reagent (Qiagen, Netherland) was applied for the transfection according to the manufacturer’s instructions.
Cell Proliferation Assay
Cells were seeded into 96-well plates and cultured overnight at 37 °C. After transfection with siTIMP1 and the corresponding siControl for 48 h, 10 µL CCK-8 solution was added into each well. After an additional incubation at 37 °C for 2 h, the absorbance was measured at 450 nm using the microplate reader (Dynatech, USA).
Scratch Wound Assay
The scratch wound assay was employed to measure the cell movement. RKO cells were cultured in 6-well plate, and the wound was achieved using a 0.1 µL pipette tip. Images were captured after transfection with siTIMP1 and the corresponding siControl for 48 h to determine the wound closure rate.
Quantitative Reverse Transcription Polymerase Chain Reaction (Qrt-PCR)
RNA was extracted by using the Trizol reagent (Invitrogen, USA). The cDNA was synthesized by using the PrimeScript™ RT reagent kit (Takara, Japan). qRT‑PCR was performed by the ABI 7900HT Real-time PCR System (Applied Biosystem, USA) using the SYBR® Green mix (Takara, Japan). Data were analyzed with the comparative Ct (ΔΔCt) method to assess the relative expression, and the GAPDH was employed as the internal control as previously introduced.26–29 The primers for GAPDH and TIMP1 were obtained from the Sangon Biological Engineering (Shanghai, China).
Apoptosis Assay
After transfection with siRNA, cells were collected, washed with PBS, and stained using the Annexin V-FITC/PI Apoptosis kit (Beyotime, China) according to the manufacturer’s instructions. Stained cells were analyzed with the BD FACSAria III flow cytometer.
Statistical Analysis
Data were shown as mean ± standard deviation (SD). Two groups were compared by using the two-tailed Student’s t-test. A significant difference was considered as p < 0.05. GraphPad Prism 7 (GraphPad Software, Inc., USA) was used to analyze all the data.
Results
Functional Characterization Of DEGs Significantly Upregulated In UC And CRC
To obtain the genes that function importantly during the initiation and progression of UC-associated CRC (ucaCRC), we first retrieved the GSE9452 dataset and obtained 568 significantly DEGs in the UC samples (Figure 1A). Among these DEGs, 426 genes were significantly upregulated, and 142 genes were significantly downregulated in UC (Figure 1B). Next, the GSE8671 dataset containing 32 CRC tissues and the corresponding normal mucosa from the same individuals was also retrieved, and 3154 significantly DEGs were identified (Figure 1C). Among these DEGs, 930 genes were significantly upregulated, and 2224 genes were significantly downregulated in CRC (Figure 1D). Considering the genes consistently upregulated in both UC and CRC are more likely to be critical for the malignant transformation,20 we compared the genes that were significantly upregulated in UC and CRC and eventually obtained 78 communal candidates (Figure 1E). To explore the functional enrichments of the 78 genes, GO and KEGG analyses were applied as reported previously.30 As shown in Figure 1F, these genes were highly enriched in the response to wounding (biological process), the extracellular region (cellular component), and the endopeptidase activity (molecular function). Additionally, these genes were also enriched in the complement and coagulation cascades, chemokine, and NOD-like receptor signaling pathways (Figure 1G). Altogether, our results screen out 78 genes that may function importantly in the initiation and progression of ucaCRC.
TIMP1 Is Identified To Be Consistently Upregulated In The Initiation And Progression Of ucaCRC
Receiving a regular intake of mesalazine has been validated to reduce the risk of ucaCRC through altering the expression of several genes, suggesting that these target genes may function importantly in the progression of ucaCRC.31,32 Additionally, increasing evidences have pointed out that inter- or intracellular proteins interactions can facilitate cancer progression.33,34 Therefore, on the one hand, identifying the candidates and their potential interacting genes based on the comprehensive bioinformatics analyses would be more helpful in recognizing the critical oncogenes of ucaCRC. On the other hand, these genes upregulated constantly during the progression of ucaCRC and simultaneously showing interaction with the target genes of mesalazine are more likely to be the potential therapeutic targets. Collectively, to obtain the potential genes that are important in the initiation and progression of ucaCRC, we made assumption that the genes should be consistently upregulated during the malignant transformation process and also the effective target for therapeutic drugs in both diseases. As a result, we first acquired 7 target genes of mesalazine including ALOX5, CHUK, IKBKB, MPO, PPARG, PTGS1, and PTGS2 (Figure 2A). The PPI network was subsequently constructed based on the 78 identified genes (Figure 1E) and these 7 target genes. The module with highest scores was generated via using the plug-ins MCODE in Cytoscape, and 8 hub genes were obtained (Figure 2B). To explore whether these hub genes were also effective targets for mesalazine in UC and CRC, we determined the mRNA expression of these genes after mesalazine treatment by retrieving the GSE46451 and GSE14538 datasets. As shown in Figure 2C, among these hub genes, the expression of TIMP1 was found to be remarkably reduced in UC and CRC after mesalazine treatment suggesting that TIMP1 is a therapeutic target for mesalazine. It has been established that the malignant transformation of ucaCRC commonly involves “inflammation-dysplasia-cancer” (Figure 2D), and DSS induced-ucaCRC is a canonical model to study this malignant process.35 Therefore, to examine the expression pattern of TIMP1 in this whole pathophysiological process, we retrieved the GSE31106 dataset and found it was consistently upregulated (Figure 2E). Taken together, these results suggest that TIMP1 may be important in the initiation and progression of ucaCRC.
Identification Of TIMP1 As A Potential Biomarker For The Worse Prognosis Of Patients With CRC
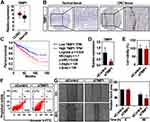
To validate the expression of TIMP1 in CRC, we further employed the GEPIA, a bioinformatics website based on TCGA and GTEx data,36 and found that the expression of TIMP1 is higher in CRC compared to normal colorectal tissues (Figure 3A). Because the expression of genes may be inconsistent with their protein level,37 we then validated the protein level of TIMP1 in CRC tissues. By using the Human Protein Atlas database, we found TIMP1 was highly expressed in the CRC tissue, but not detected in the non-malignant colorectal tissue (Figure 3B). To assess the association between the TIMP1 expression and the clinical outcome of patients with CRC, as shown in Figure 3C, GEPIA results revealed the expression of TIMP1 was negatively associated with the overall survival in patients with CRC (P = 0.036). To explore the role of TIMP1 in the proliferation and movement of CRC in vitro, we then decreased the expression of TIMP1 in RKO cells (Figure 3D). The cell proliferation and apoptosis rate of RKO cells were not significantly altered by TIMP1 knockdown (Figure 3E and F). However, interestingly, knockdown of TIMP1 significantly increased the cell migration of RKO cells suggesting that TIMP1 may function as an inhibitor of cell movement in vitro (Figure 3G). Collectively, our data suggest that upregulated TIMP1 can be a potential biomarker for the prognosis of patients with CRC and also functions importantly in the cancer cell movement in vitro.
Mechanistical Validation Of TIMP1 In Driving The Malignant Transformation Of ucaCRC
To mechanistically verify that TIMP1 functions importantly in the ucaCRC, the GeneMANIA was used. As shown in Figure 4A, 20 proteins/genes might interact with TIMP1, and 3 genes including MMP9, STAT3, and JUN have been studied to be participated in the malignant transformation from to UC to CRC.38–40 As shown in Figure 4B, most of these genes showed no significantly alteration in UC and CRC except for MMP9 in UC. Identification of the transcription factors regulating TIMP1 can be valuable in elucidating the underlying mechanisms of TIMP1-mediated ucaCRC.14 We therefore employed the Funrich to predict the potential transcription factors, and HOXA9, PLAU, ATF1, KLF7, and SP1 were selected to be the top 5 candidates (Figure 4C). Among the factors, SP1 has been reported to be functional in promoting ucaCRC.41 The Coremine Medical database was applied to annotate the enriched biological processes of TIMP1. As shown in Figure 4D, there were ten biological processes were closely associated with the pathological processes, and TIMP1 could participate in such process by altering the cell proliferation, the cell cycle and also the cell migration. Therefore, these results indicate that TIMP1 may function importantly in the ucaCRC through various mechanisms.
Discussion
Herein, TIMP1 is identified to be consistently upregulated in the initiation and progression of ucaCRC and can also be a potential biomarker for the prognosis of patients with CRC based on the integrative bioinformatics analyses. The role of TIMP1 in the malignant transformation of ucaCRC is further validated via employing several online tools including the protein/gene interactions, the biological process annotation, and the transcription factors identification.
The pathogenesis of CRC in patients with UC is largely distinct from that of sporadic CRC and has been recognized as a step-wise progression from inflamed epithelia to dysplasia and eventually cancer. Malignant transformation is one of the most serious complications of UC, in which the homeostasis of the intestinal immune system is severely disrupted.42 The excessive reactive oxygen species generated by chronic inflammation in the colorectal mucosa can be the main cause of DNA damage in patients with UC that eventually leads to CRC.43 There are two major options provided currently for CRC screening in patients with UC including the more frequent surveillance and the colectomy based on the degree of dysplasia examined by the colonoscopy.44 However, dysplasia-based screening is comparatively problematic and has a poor success rate in terms of identifying patients who are likely to turn into cancer. Additionally, there is also a high degree of intraobserver variability regarding the interpretation of histological images.45 As a result, novel and reliable biomarkers for premalignancy screening are urgently required. Herein, TIMP1 is identified as a pivotal candidate in promoting the development of ucaCRC. Mutations in p53 and adenomatous polyposis coli have long been revealed as critical events for ucaCRC,46,47 and the mutation detection based on tissues biopsy have also yielded promising results as biomarkers of premalignancy in patients with UC.48 As the level of TIMP1 has been suggested to be associated with the prognosis of several cancer types such as breast and pancreatic cancer,49–51 and given that the expression of TIMP1 is consistently upregulated during this malignant transformation (Figure 2E) and negatively correlates with the prognosis of CRC patients (Figure 3C), we thus conclude that TIMP1 can be also a potential biomarker for CRC screening in patients with UC.
TIMPs are dimers consisting of a smaller C-terminal domain and an N-terminal domain binding to the MMPs substrate and thereby essential for MMPs degradation.52 TIMP1, the inducible form, belongs to a glycoprotein group with four members (TIMP1-4), whose main function is to mediate extracellular matrix turnover. TIMP1 has been demonstrated to participate in multiple pathological processes such as wound healing and cancer metastasis.52 Consistent with these previous findings, our results indicate that TIMP1 can function as an intracellular inhibitor in the cancer cells movement (Figure 3G), which can be closely related with the expression or function of MMP9 (Figure 4A). Breakdown of extracellular matrix mediated by MMPs is necessary for the migration of cancer or immune cells during metastasis.53 Previous studies have pointed out that MMP9 is significantly upregulated in the DSS-induced UC, and targeting of MMP9 can relieve the experimental symptoms of UC.54,55 TIMP1 has also been reported to inhibit macrophages angiogenic activity through forming specific form with MMP9.56 Based on these findings, we therefore speculate that the interaction between TIMP1 and MMP9 may function importantly in the initiation and progression in sporadic CRC and ucaCRC.
Conclusion
Collectively, our findings suggest that TIMP1 consistently upregulated in the pathological process of ucaCRC can also be a potential biomarker for the worse prognosis of CRC. Uncovering this novel role of TIMP1 in the malignant transformation may extend our understanding of the pathogenesis from normal mucosa to ucaCRC.
Acknowledgment
This research was funded by the National Natural Science Foundation of China (Grant numbers: 81573004, 81773275, and 81871953).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Menon M, Cunningham C, Kerr D. Addressing unwarranted variations in colorectal cancer outcomes: a conceptual approach. Nat Rev Clin Oncol. 2016;13(11):706–712. doi:10.1038/nrclinonc.2016.94
3. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi:10.1056/NEJMra0804588
4. Garrett WS, Punit S, Gallini CA, et al. Colitis-associated colorectal cancer driven by T-bet deficiency in dendritic cells. Cancer Cell. 2009;16(3):208–219. doi:10.1016/j.ccr.2009.07.015
5. Grivennikov S, Karin E, Terzic J, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15(2):103–113. doi:10.1016/j.ccr.2009.01.001
6. Bernstein CN, Blanchard JF, Kliewer E, Wajda A. Cancer risk in patients with inflammatory bowel disease: a population-based study. Cancer. 2001;91(4):854–862. doi:10.1002/(ISSN)1097-0142
7. Pekow J, Meckel K, Dougherty U, et al. miR-193a-3p is a key tumor suppressor in ulcerative colitis-associated colon cancer and promotes carcinogenesis through upregulation of IL17RD. Clin Cancer Res. 2017;23(17):5281–5291. doi:10.1158/1078-0432.CCR-17-0171
8. Han J, Jackson D, Holm J, et al. Elevated d-2-hydroxyglutarate during colitis drives progression to colorectal cancer. Proc Natl Acad Sci U S A. 2018;115(5):1057–1062. doi:10.1073/pnas.1712625115
9. Ungaro R, Mehandru S, Allen PB, Peyrin-Biroulet L, Colombel JF. Ulcerative colitis. Lancet (London, England). 2017;389(10080):1756–1770. doi:10.1016/S0140-6736(16)32126-2
10. Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5(10):588–599. doi:10.1038/ncponc1187
11. Ioannidis JP, Allison DB, Ball CA, et al. Repeatability of published microarray gene expression analyses. Nat Genet. 2009;41(2):149–155.
12. Lee H, Palm J, Grimes SM, Ji HP. The cancer genome atlas clinical explorer: a web and mobile interface for identifying clinical-genomic driver associations. Genome Med. 2015;7:112. doi:10.1186/s13073-015-0226-3
13. Huang R, Gao L. Identification of potential diagnostic and prognostic biomarkers in non-small cell lung cancer based on microarray data. Oncol Lett. 2018;15(5):6436–6442. doi:10.3892/ol.2018.8153
14. You S, Gao L. Identification of NMU as a potential gene conferring alectinib resistance in non-small cell lung cancer based on bioinformatics analyses. Gene. 2018;678:137–142. doi:10.1016/j.gene.2018.08.032
15. Huang R, Gu W, Sun B, Gao L. Identification of COL4A1 as a potential gene conferring trastuzumab resistance in gastric cancer based on bioinformatics analysis. Mol Med Rep. 2018;17(5):6387–6396. doi:10.3892/mmr.2018.8664
16. Olsen J, Gerds TA, Seidelin JB, et al. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis. 2009;15(7):1032–1038. doi:10.1002/ibd.20879
17. Sabates-Bellver J, Van der Flier LG, de Palo M, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5(12):1263–1275. doi:10.1158/1541-7786.MCR-07-0267
18. Parenti S, Ferrarini F, Zini R, et al. Mesalazine inhibits the beta-catenin signalling pathway acting through the upregulation of mu-protocadherin gene in colo-rectal cancer cells. Aliment Pharmacol Ther. 2010;31(1):108–119. doi:10.1111/j.1365-2036.2009.04149.x
19. MacFie TS, Poulsom R, Parker A, et al. DUOX2 and DUOXA2 form the predominant enzyme system capable of producing the reactive oxygen species H2O2 in active ulcerative colitis and are modulated by 5-aminosalicylic acid. Inflamm Bowel Dis. 2014;20(3):514–524. doi:10.1097/01.MIB.0000442012.45038.0e
20. Tang A, Li N, Li X, et al. Dynamic activation of the key pathways: linking colitis to colorectal cancer in a mouse model. Carcinogenesis. 2012;33(7):1375–1383. doi:10.1093/carcin/bgs183
21. Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi:10.1093/nar/gks1193
22. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57.
23. Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi:10.1093/nar/gku1003
24. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(Web Server issue):W214–W220. doi:10.1093/nar/gkq537
25. Benito-Martin A, Peinado H. FunRich proteomics software analysis, let the fun begin! Proteomics. 2015;15(15):2555–2556. doi:10.1002/pmic.v15.15
26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego, Calif). 2001;25(4):402–408. doi:10.1006/meth.2001.1262
27. Popivanova BK, Kostadinova FI, Furuichi K, et al. Blockade of a chemokine, CCL2, reduces chronic colitis-associated carcinogenesis in mice. Cancer Res. 2009;69(19):7884–7892. doi:10.1158/0008-5472.CAN-09-1451
28. Polytarchou C, Hommes DW, Palumbo T, et al. MicroRNA214 is associated with progression of ulcerative colitis, and inhibition reduces development of colitis and colitis-associated cancer in mice. Gastroenterology. 2015;149(4):981–992.e911. doi:10.1053/j.gastro.2015.05.057
29. Nakatsuji M, Minami M, Seno H, et al. EP4 receptor-associated protein in macrophages ameliorates colitis and colitis-associated tumorigenesis. PLoS Genet. 2015;11(10):e1005542. doi:10.1371/journal.pgen.1005542
30. Xing Z, Chu C, Chen L, Kong X. The use of gene ontology terms and KEGG pathways for analysis and prediction of oncogenes. Biochim Biophys Acta. 2016;1860(11 Pt B):2725–2734. doi:10.1016/j.bbagen.2016.01.012
31. Bonovas S, Fiorino G, Lytras T, Nikolopoulos G, Peyrin-Biroulet L, Danese S. Systematic review with meta-analysis: use of 5-aminosalicylates and risk of colorectal neoplasia in patients with inflammatory bowel disease. Aliment Pharmacol Ther. 2017;45(9):1179–1192. doi:10.1111/apt.14023
32. van Staa TP, Card T, Logan RF, Leufkens HG. 5-Aminosalicylate use and colorectal cancer risk in inflammatory bowel disease: a large epidemiological study. Gut. 2005;54(11):1573–1578. doi:10.1136/gut.2005.070896
33. McElhinny AS, Li JL, Wu L. Mastermind-like transcriptional co-activators: emerging roles in regulating cross talk among multiple signaling pathways. Oncogene. 2008;27(38):5138–5147. doi:10.1038/onc.2008.228
34. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
35. Zisman TL, Rubin DT. Colorectal cancer and dysplasia in inflammatory bowel disease. World J Gastroenterol. 2008;14(17):2662–2669. doi:10.3748/wjg.14.2662
36. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi:10.1093/nar/gkx247
37. Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583(24):3966–3973. doi:10.1016/j.febslet.2009.10.036
38. Li Y, de Haar C, Chen M, et al. Disease-related expression of the IL6/STAT3/SOCS3 signalling pathway in ulcerative colitis and ulcerative colitis-related carcinogenesis. Gut. 2010;59(2):227–235. doi:10.1136/gut.2009.184176
39. Thorsteinsdottir S, Gudjonsson T, Nielsen OH, Vainer B, Seidelin JB. Pathogenesis and biomarkers of carcinogenesis in ulcerative colitis. Nat Rev Gastroenterol Hepatol. 2011;8(7):395–404. doi:10.1038/nrgastro.2011.96
40. Marshall DC, Lyman SK, McCauley S, et al. Selective allosteric inhibition of MMP9 is efficacious in preclinical models of ulcerative colitis and colorectal cancer. PLoS One. 2015;10(5):e0127063. doi:10.1371/journal.pone.0127063
41. Pugachev KK, Shimbireva IB, Belous TA, Frank GA. [The immunohistochemical study of trophoblast beta 1-globulin expression in cancer and in the mucosa of the large intestine]. Biull Eksp Biol Med. 1992;114(11):513–515.
42. Rutter M, Saunders B, Wilkinson K, et al. Severity of inflammation is a risk factor for colorectal neoplasia in ulcerative colitis. Gastroenterology. 2004;126(2):451–459. doi:10.1053/j.gastro.2003.11.010
43. Lopez A, Pouillon L, Beaugerie L, Danese S, Peyrin-Biroulet L. Colorectal cancer prevention in patients with ulcerative colitis. Best Pract Res Clin Gastroenterol. 2018;32–33:103–109. doi:10.1016/j.bpg.2018.05.010
44. Fujii S, Katsumata D, Fujimori T. Limits of diagnosis and molecular markers for early detection of ulcerative colitis-associated colorectal neoplasia. Digestion. 2008;77(Suppl 1):2–12. doi:10.1159/000111482
45. Chambers WM, Warren BF, Jewell DP, Mortensen NJ. Cancer surveillance in ulcerative colitis. Br J Surg. 2005;92(8):928–936. doi:10.1002/(ISSN)1365-2168
46. Redston MS, Papadopoulos N, Caldas C, Kinzler KW, Kern SE. Common occurrence of APC and K-ras gene mutations in the spectrum of colitis-associated neoplasias. Gastroenterology. 1995;108(2):383–392. doi:10.1016/0016-5085(95)90064-0
47. Hussain SP, Amstad P, Raja K, et al. Increased p53 mutation load in noncancerous colon tissue from ulcerative colitis: a cancer-prone chronic inflammatory disease. Cancer Res. 2000;60(13):3333–3337.
48. Willenbucher RF, Zelman SJ, Ferrell LD, Moore DH
49. Schrohl AS, Holten-Andersen MN, Peters HA, et al. Tumor tissue levels of tissue inhibitor of metalloproteinase-1 as a prognostic marker in primary breast cancer. Clin Cancer Res. 2004;10(7):2289–2298. doi:10.1158/1078-0432.CCR-03-0360
50. Grunnet M, Mau-Sorensen M, Brunner N. Tissue inhibitor of metalloproteinase 1 (TIMP-1) as a biomarker in gastric cancer: a review. Scand J Gastroenterol. 2013;48(8):899–905. doi:10.3109/00365521.2013.812235
51. Roy R, Zurakowski D, Wischhusen J, et al. Urinary TIMP-1 and MMP-2 levels detect the presence of pancreatic malignancies. Br J Cancer. 2014;111(9):1772–1779. doi:10.1038/bjc.2014.462
52. Gardner J, Ghorpade A. Tissue inhibitor of metalloproteinase (TIMP)-1: the TIMPed balance of matrix metalloproteinases in the central nervous system. J Neurosci Res. 2003;74(6):801–806. doi:10.1002/(ISSN)1097-4547
53. Murphy G, Nagase H. Progress in matrix metalloproteinase research. Mol Aspects Med. 2008;29(5):290–308. doi:10.1016/j.mam.2008.05.002
54. Castaneda FE, Walia B, Vijay-Kumar M, et al. Targeted deletion of metalloproteinase 9 attenuates experimental colitis in mice: central role of epithelial-derived MMP. Gastroenterology. 2005;129(6):1991–2008. doi:10.1053/j.gastro.2005.09.017
55. Ishida K, Takai S, Murano M, et al. Role of chymase-dependent matrix metalloproteinase-9 activation in mice with dextran sodium sulfate-induced colitis. J Pharmacol Exp Ther. 2008;324(2):422–426. doi:10.1124/jpet.107.131946
56. Mook OR, Frederiks WM, Van Noorden CJ. The role of gelatinases in colorectal cancer progression and metastasis. Biochim Biophys Acta. 2004;1705(2):69–89. doi:10.1016/j.bbcan.2004.09.006
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.