Back to Journals » International Journal of Nanomedicine » Volume 16
Therapeutic Potential of Exosomal circRNA Derived from Synovial Mesenchymal Cells via Targeting circEDIL3/miR-485-3p/PIAS3/STAT3/VEGF Functional Module in Rheumatoid Arthritis
Authors Zhang J, Zhang Y, Ma Y, Luo L, Chu M, Zhang Z
Received 19 August 2021
Accepted for publication 23 November 2021
Published 3 December 2021 Volume 2021:16 Pages 7977—7994
DOI https://doi.org/10.2147/IJN.S333465
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Mian Wang
Juan Zhang,1 Yue Zhang,1 Yeye Ma,1 Lili Luo,1 Maolin Chu,2 Zhiyi Zhang1
1Department of Rheumatology, The First Affiliated Hospital, Harbin Medical University, Harbin, People’s Republic of China; 2Department of Urology, The Second Affiliated Hospital, Harbin Medical University, Harbin, People’s Republic of China
Correspondence: Zhiyi Zhang; Maolin Chu Tel +86 13303608999
; +86 451 86605579
Fax +86 451 85556421
Email [email protected]; [email protected]
Background: Synovial inflammation and its associated activation of angiogenesis play critical roles in rheumatoid arthritis (RA). Exosomes, as carriers of genetic information including circular RNAs (circRNAs), have been explored as delivery vehicles for therapeutic molecules. However, the effects of synovial mesenchymal stem cells (SMSCs)-derived exosomal circRNAs and their mechanisms of action in RA progression remain unclear.
Methods: SMSCs-derived exosomes (SMSCs-Exos) were administered to a co-culture of RA fibroblast-like synoviocytes (RA-FLS) and human dermal microvascular endothelial cells (HDMECs) in vitro as well as to a collagen-induced arthritis (CIA) mouse model in vivo. Their effects on VEGF expression and angiogenic activity in vitro and the therapeutic efficacy in vivo were evaluated. Exosomes from circEDIL3-overexpressing SMSCs (Ad-circEDIL3-SMSCs-Exos) were used to further determine the role of circEDIL3 in SMSCs-Exo-based therapy.
Results: Both SMSCs-Exos and Ad-circEDIL3-SMSCs-Exos significantly downregulated the expression of VEGF induced by the IL-6/sIL-6R complex in the supernatants of RA-FLS and HDMECs co-culture as well as in the cell lysate of co-cultured RA-FLS, and the extent of reduction was more pronounced in the latter. Subsequent experiments showed that angiogenic activity was significantly downregulated by SMSCs-Exos and Ad-circEDIL3-SMSCs-Exos due to reduced VEGF expression. CircEDIL3 functioned as a sponge for miR-485-3p, which targeted PIAS3. PIAS3 is known to suppress STAT3 activity and reduce downstream VEGF. Injection of SMSCs-Exos or Ad-circEDIL3-SMSCs-Exos reduced synovial VEGF and consequently ameliorated arthritis severity in the CIA mouse model.
Conclusion: The intracellular transfer of circEDIL3 by SMSCs-Exos may be a potential novel therapeutic strategy for RA.
Keywords: circEDIL3, exosome, PIAS3, STAT3, VEGF
Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory disease characterized by synovial pannus formation resulting from angiogenesis accompanied by inflammation, which leads to cartilage and bone destruction.1 The inflammatory state in RA is maintained by the transport of inflammatory cells and oxygen to the synovium through enhanced angiogenesis. Therefore, angiogenesis has been explored as a promising therapeutic target for the treatment of RA. The synovium of RA patients contains fibroblast-like synoviocytes (FLSs), which copiously produce angiogenic factors such as vascular endothelial growth factor (VEGF). Wang et al and Li et al verified that dysregulated autophagy2 and dysimmunity3 are involved in the progression of RA synovial pannus formation. In particular, we found that the angiogenic functional module, of which VEGF is the key member, played a critically important role in angiogenesis, suggesting that agents that inhibit VEGF production could be applied as antirheumatic drugs.4 However, the current therapy with monoclonal antibodies targeting VEGF is limited in clinical application due to adverse reactions such as hypertension and thromboembolism. Therefore, therapeutic targets more specific than VEGF are needed.5
Circular RNAs (circRNAs), a type of noncoding RNA, represent a novel class of abundant, widespread and tissue-specific transcripts that form a covalently closed continuous loop.6 CircRNAs regulate gene expression primarily by functioning as efficient sponges of microRNAs (miRNAs), which can bind to complementary sequences in the 3ʹ untranslated region (3ʹ-UTR) of target mRNAs and either repress translation or degrade targets.7 CircRNAs have been reported to be involved in the pathogenesis of diseases including tumours;8 however, due to the tissue specificity, the role of circRNAs in RA remains unclear, and there has been no report on circRNA that might play a key functional role in both angiogenesis and inflammation. In fact, in various diseases, inflammation and angiogenesis often follow and promote each other, and play critical roles in the initiation and progression of diseases.9 A systematic study of circRNAs and their influence on neovascularization as it relates to inflammation is necessary to develop them for clinical application. Additionally, despite the potential of circRNAs as therapeutic agents, an effective method for their delivery to the target tissue is still lacking.
Exosomes (vesicles ranging from 30–200 nm in size) have gained increasing interest as delivery vehicles for the transfer of noncoding RNAs.10,11 A study showed that exosomes released from mesenchymal stem cells (MSCs) could protect T lymphocytes from inflammation12 and that MSC-derived exosomes could reduce joint destruction by inhibiting synoviocyte hyperplasia and angiogenesis.13 Although circRNAs are enriched and stable in exosomes,11 biological functions of circRNAs secreted from MSC-derived exosomes in inflammation and angiogenesis remain undefined. To fill this knowledge gap, we designed the current study with the aim of investigating the regulatory function of MSC-derived exosomal circRNAs in the inflammation-induced angiogenic response in RA.
In the present study, one of the circRNAs (hsa_circ_0073244, circEDIL3) was found to be significantly expressed in normal synovial MSCs (SMSCs). Under inflammatory conditions induced by IL-6, SMSCs-derived exosomal circEDIL3 reduced the levels of VEGF secreted by RA-FLS in a dose-dependent manner. Using an online tool to predict miRNA-mRNA interaction, we found that circEDIL3 targeted miR-485-3p, which is predicted to target Protein Inhibitor of Activated STAT3 (signal transducer and activator of transcription 3) [PIAS3]. PIAS3 belongs to the small Rho GTPase family and is the main cellular inhibitor of STAT3. Strikingly, our experiment showed that exosomal circEDIL3 derived from SMSCs could target the VEGF functional module via the miR-485-3p/PIAS3/STAT3 axis in a CIA model. Hence, circRNAs may be a new class of therapeutics for RA, and secreted circEDIL3 can be transferred into RA-FLS with intact functional activity by the novel bioengineered carrier SMSCs-Exos. This new system might be instrumental in alleviating disease progression by mitigating inflammation-induced angiogenesis. It holds great therapeutic potential for RA and possibly for other diseases.
Materials and Methods
Isolation, Culture, and Differentiation of SMSCs
Synovial MSCs (SMSCs) were isolated and expanded as previously reported.14 Briefly, synovial tissue from human donors of various ages (mean 44 years, range 30–66 years) was obtained at arthroscopic surgeries of knee joints in accordance with a protocol approved by the Ethics Committee of the First Affiliated Hospital of Harbin Medical University, and signed consents were obtained from the donors. Synovium was digested with collagenase (Worthington Biochemical, Lakewood, NJ, USA) overnight at 37 °C, and the cells were collected and cultured in high-glucose DMEM supplemented with 10% FBS (HyClone, Logan, UT, USA). Cells obtained after 14 days were designated at Passage 0. After an additional 14 days, single-cell-derived colonies were obtained and pooled together. The cells were identified as SMSCs by flow cytometry (FCM) with Abs against CD105, CD44, CD90, CD73, CD11b, CD14 and CD34 (BD Biosciences). We further confirmed MSCs by adipogenesis, osteogenesis and chondrogenesis assays using specific differentiation medium (CTCC, Wuxi, China). Cells at passages 4–7 were used in the present studies.
Isolation and Confirmation of Exosomes from SMSC Culture Medium
Exosomes in SMSC culture medium were isolated by differential centrifugation according to previous reports.15 Briefly, after incubating SMSCs in exosome-free cell growth medium (SBI System Biosciences) for 48 h, the culture medium was centrifuged at 3000× g for 30 min to remove cell debris, and the supernatant was collected and centrifuged at 16,500 × g for 30 min. Finally, the supernatant was centrifuged at 120,000 × g for 120 min, and exosomes were harvested and stored at −80 °C.
For conventional transmission electron microscopy (TEM), exosome pellets were dropped in a carbon-coated copper grid, stained with 2% uranyl acetate, and observed with a transmission electron microscope (Libra 200 FE, Zeiss, Germany). Exosomes were also identified by nanoparticle tracking analysis (NTA) using a Zetasizer Nano ZS (Malvern, UK) and Western blot analysis.
Incubation of SMSCs-Exos with the Co-Culture of RA-FLS and HDMECs
RA fibroblast-like synoviocytes (RA-FLS) were purchased from Cell Applications (San Diego, CA, USA) and maintained in 10% FBS DMEM. Human dermal microvascular endothelial cells (HDMECs) were obtained from ScienCell Research Laboratories (6076 Corte Del Cedro, Carlsbad, CA, USA) and cultured in Endothelial Cell Growth Medium (EGM)-2 from the same company. Cells at passages 3–6 were used for experiments. A co-culture system of RA-FLS and HDMECs was established as described previously.4 Briefly, RA-FLS (4×105 cells/mL) and HDMECs (ranging from 2×105 cells/mL to 3×105 cells/mL) were seeded in the lower and upper chambers of a Transwell apparatus (Costar), respectively, and incubated in exosome-free cell growth medium (SBI). The culture was stimulated with IL-6/sIL-6R complex (IL-6100 ng/mL, sIL-6R 100 ng/mL) as previously described.16 Exosomes from SMSCs (SMSCs-Exos, 100 µg) were then added to the co-culture. After incubating in 1% FBS DMEM for 48 h, the supernatants were harvested and centrifuged at 1000 rpm to remove cellular debris and further centrifuged at 120,000 × g for 120 min to remove exosomes. A portion of the culture supernatants were removed for the subsequent Transwell assay, tube formation test and ex vivo aortic ring angiogenesis assay. The remaining supernatants were frozen at −20°C until analysis with a commercial ELISA kit for VEGF (R&D Systems). The mRNA and protein expression in co-cultured RA-FLS were determined by quantitative real-time PCR (qRT-PCR) and Western blot, respectively.
HDMEC Transwell Assay, in vitro Tube Formation Assay, and ex vivo Aortic Ring Angiogenesis Assay
The Transwell assay and the aortic ring assay were performed as described previously.4 Briefly, for the Transwell assay, 600 µL of the supernatants from the co-cultures were placed in the lower chamber. HDMECs (3×104 cells) were added to the upper chamber and incubated for 6 h. Cells that migrated to the bottom of the Transwell membrane were stained and quantified (LEICA DMi8). Aortic rings isolated from 8-week-old C57BL/6 mice (SPF, Laboratory Animal Center of the Second Affiliated Hospital of Harbin Medical University) were incubated for 3 days in 1% FBS EGM-2 on Matrigel. After verifying sprouts from the aortic rings, EGM-2 was replaced with 500 μL of fresh supernatants from the co-culture of RA-FLS and HDMECs, and the aortic rings were incubated for additional 3 days. Microvessel outgrowths were examined with a microscope (LEICA DMi8) and analysed using ImageJ software. In the tube formation assay, a µ-slide (ibidi, GmbH, Munich, Germany) was used to investigate angiogenesis. Each well was coated with 10 μL of Matrigel (Corning, New York, USA), and 70 μL of cell suspension containing 1×104 cells was added to each well. After 6 h of incubation, endothelial cell tube formation was assessed using phase-contrast microscopy (LEICA DMi8).
Exosome Labelling
The PKH67 Green Fluorescent Cell Linker Kit (Sigma) was used for exosome staining. Exosomes were resuspended in 100 μL of diluent C, mixed with 100 μL PKH67 dye solution (4×10−6 M), and incubated for 1–5 min. The reaction was stopped by adding 200 μL of exosome-free serum. The labelled exosomes were then washed twice with PBS by centrifugation (100,000×g for 1 h) and incubated with recipient RA-FLS for 6 h before imaging was performed under a laser confocal microscope (LEICA TCS SP8).
RNA Isolation and qRT-PCR
Total RNA was extracted from cultured cells with TRIzol reagent (Invitrogen) as described before.17 For exosome-RNA isolation, Acryl Carrier (Solarbio, Beijing, China) and isopropanol were added after the upper water phase was transferred to the new EP tube. For circRNA detection, total RNA was incubated with RNase R (Geneseed Biotech, Guangzhou, China). cDNA for mRNA and circRNA analyses was synthesized with PrimeScript RT Master Mix (Takara Biotechnology, Dalian, China), and qRT–PCR amplification was performed using iTaq Universal SYBR Green Supermix (Bio–Rad). For miRNA detection, cDNA synthesis was performed using a riboSCRIPTTM Reverse Transcription Kit (RiboBio, Guangzhou, China), and qRT–PCR was performed with an Applied Biosystems TaqMan MicroRNA Assay Kit. Relative circRNA, mRNA or miRNA expression was normalized to GAPDH or U6 snRNA levels using the 2−ΔΔCt method. Experiments were performed in triplicate for each sample. The sequence for each primer is listed in Supplementary Table 1.
RNA FISH
Fluorescence in situ hybridization (FISH) was performed with Fluorescent In situ Hybridization Kits (RiboBio, Guangzhou, China). Briefly, after treatment in pre-hybridization buffer, RA-FLS grown on coverslips were incubated in hybridization buffer for 12–16 h with a CY3-labelled circRNA probe (RiboBio, Guangzhou, China, Supplementary Table 1) in a humid and dark environment. The nuclei were stained with DAPI, and intracellular localization of circRNA was observed using a TCS SP8 X laser confocal microscope (LEICA).
Bioinformatics Analysis
The miRNA target of circEDIL3 was predicted by the bioinformatics databases TargetScan (http://www.targetscan.org/) and CircInteractome (https://circinteractome.nia.nih.gov/). The mRNA target of miR-485-3p was predicted by TargetScan (http://www.targetscan.org/) and Starbase (http://starbase.sysu.edu.cn/).
Virus Infection and Cell Transfection
Adenovirus vectors (Ad-Vectors) encoding circEDIL3 (Ad-circEDIL3), PIAS3 (Ad-PIAS3), STAT3 (Ad-STAT3) and GFP control were prepared by Hanbio Biotechnology (Shanghai, China). In brief, circEDIL3, PIAS3 and STAT3 fragments were amplified and ligated with the linearized vector, and the amplified sequences were verified by Sanger sequencing. Packaging plasmids and viral vectors were co-transfected into HEK-293T cells using Lipofectamine 3000 transfection reagent (Thermo Fisher). Forty-eight hours after transfection, the culture medium was centrifuged and supplemented with polybrene (SolarBio). Finally, the culture medium mixed with polybrene was added to target cells for infection. CircEDIL3 overexpression was established in SMSCs and RA-FLS. PIAS3 and STAT3 overexpression was established in RA-FLS.
A predesigned siRNA targeting human circEDIL3 (si-circEDIL3) was produced by GenePharma (Shanghai, China, Supplementary Table S1) and transfected into RA-FLS using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Cells were grown for 48 h post-transfection and harvested. The vectors that upregulate or downregulate relative miRNA expression (miRNA mimics and miRNA inhibitors) were designed and constructed by RiboBio (Guangzhou, China).
RNA Pull-Down Assay
For the biotin-coupled circRNA pull-down assay, the probe was designed to bind to the junction area of circEDIL3 and synthesized by GenePharma (Shanghai, China). RA-FLS were washed in ice-cold PBS, lysed in lysis buffer and incubated with 3 µg biotinylated probes against the circEDIL3 sequence or negative control. The cell lysates with biotinylated probes were incubated with 50 µL streptavidin-conjugated magnetic beads (Invitrogen, USA), and TRIzol reagent (Invitrogen, Thermo, USA) was added to the washed beads for RNA extraction. RNAs were reverse transcribed and analysed by qRT–PCR assay.
For biotin-coupled miRNA capture, 3′-end biotinylated miR-485-3p mimics or control RNA (RiboBio, Guangzhou, China) were transfected into RA-FLS at a final concentration of 50 nM. Forty-eight hours later, cells were harvested, and the cell pellet was lysed in lysis buffer with complete protease inhibitor cocktail. The abundance of circEDIL3 and PIAS3 mRNA in the bound fraction pulled down by streptavidin-coated magnetic beads was evaluated by qRT–PCR analysis.
Luciferase Reporter Assay
The expression plasmid of circEDIL3 was generated by inserting the entire human circEDIL3 sequence into the pcDNA3.1 circRNA Mini Vector (Addgene). The partial fragment of GFP was amplified by PCR and cloned into the pcDNA3.1 circRNA Mini Vector to be used as a control plasmid. RA-FLS were transfected with miR-485-3p mimic (RiboBio, China) or control mimic in combination with a luciferase reporter or an empty vector. Cells were also transfected with pcDNA3.1-circEDIL3 and its mutant. The PIAS3 gene 3ʹ-UTR containing miR-485-3p target sites or their mutant sequences was inserted into the pGL3-promoter vector. MiR-485-3p mimics/inhibitors and respective negative controls were transfected into RA-FLS at a final concentration of 100 nM using a riboFECTTM CP Kit (RiboBio, China) according to the manufacturer’s instructions. Luciferase activity was measured using a Dual-Glo Luciferase Assay System (Promega, Madison, WI) with a Cytation 5 Cell Imaging Multi-Mode Reader (BioTek) 24 h after transfection. Each sample was prepared in triplicate, and the entire experiment was repeated three times.
Western Blot Analysis
Protein extraction and Western blot analysis were performed as described before.17 Antibodies against specific proteins, including VEGF (Abcam, ab214424), PIAS3 (Cell Signaling Technology, 9042), p-STAT3 (Cell Signaling Technology, 9145), STAT3 (Cell Signaling Technology, 12640), CD63 (Abcam, ab134045) and TSG101 (Abcam, ab125011), were used.
Animal Model of Collagen-Induced Arthritis (CIA)
All animal experiments were performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals under the approval from the Institutional Animal Care and Use Committee of the First Affiliated Hospital of Harbin Medical University. Male DBA/1J mice (8 weeks old and 19 g ± 2 g in weight, SPF, SLAC, Shanghai, China) were used to establish the CIA model as described previously.4 Briefly, bovine type II collagen (CII, Chondrex) was emulsified in an equal volume of complete Freund’s adjuvant (CFA) (Chondrex). Each DBA/1J mouse was given 100 μL of the emulsion containing 100 μg CII via intradermal injection at the base of the tail. On Day 21 after primary immunization, the mice received a booster shot of 100 μg CII emulsified with an equal volume of incomplete Freund’s adjuvant (Chondrex). After induction of arthritis (on Day 21), the mice were divided into the following groups (n = 5 per experimental group): CIA control, SMSCs-Exo, Ad-circEDIL3-SMSCs-Exo, Ad-Vector-SMSCs-Exo (CIA mice treated with indicated exosomes at a dose of 100 µg suspended in 10 µL PBS twice per week), methotrexate (MTX, CIA mice treated with MTX at a dose of 1.5 mg/kg/week, as a treatment control) and a normal control group (without immunization). Intraarticular treatment lasted from Day 26 to Day 39 after the first immunization. Normal and CIA model control mice were given an equal volume of PBS. The body weights of the mice were carefully monitored every three days from Day 24 to Day 39.
Assessment of Arthritis Severity and Micro-Computed Tomography (Micro-CT) Imaging
Clinical arthritis was assessed daily from Day 21 after primary immunization, and the arthritic score was evaluated every 3 days until the end of the experimental period according to a standard scoring system.4 At the end of this study (Day 40 after the initial collagen injection), micro-CT imaging (Quantum GX, Perkin Elmer, Waltham, USA) was conducted to investigate the effects of exosomes on the three-dimensional (3D) bone structure. Mean CT values of hind paws and knee joints were calculated with Caliper Analyze software (Analyze Direct, Kansas, USA) to assess bone loss.
Histological Examination and Immunohistochemical Analysis
After euthanasia, knee joints of mice were collected and stained with haematoxylin-eosin (H&E). To quantitatively evaluate the severity of arthritis, a scoring system was employed according to the reported protocol.4 Immunohistochemical analysis was performed as previously described with some modifications.4 Knee joint sections on slides were incubated with antibodies against PIAS3, p-STAT3, STAT3 or VEGF (Wanleibio, Shenyang, China). Subsequently, the sections were stained using a polymer HRP detection system (PV9001, ZSGB-BIO, Beijing, China) and visualized with a DAB peroxidase substrate kit (ZLI-9017, ZSGB-BIO, Beijing, China). Following immunostaining, the synovial area in the joint of each section was evaluated under a microscope (LEICA DMi8, Germany) in three randomly selected areas at a magnification of 100×. Image-Pro Plus 6 (Media Cybernetics, Inc.) was used to analyse the average integrated optical density (IOD) according to a previously described protocol.2
Statistical Analysis
The data were represented as means ± S.E.M. Statistical analysis was conducted with Student’s t-test or one-way analysis of variance (ANOVA) as appropriate by using SPSS version 17.0 software. Values of p < 0.05 were considered statistically significant.
Results
SMSCs-Exo Treatment Inhibited Upregulation of RA-FLS-Induced Angiogenesis Under Inflammatory Conditions
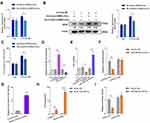
We isolated synovial MSCs (SMSCs) from humans and confirmed the MSC properties of a selected clone by a differentiation assay (Supplementary Figure 1A-C). SMSCs were also identified by flow cytometry (FCM) analysis with specific cell surface markers (Supplementary Figure 2). Exosomes from SMSCs (SMSCs-Exos) were extracted from conditioned cell medium supplemented with exosome-free FBS. Morphology and size were examined by transmission electron microscopy (TEM) (Figure 1A) and nanoparticle tracking analysis (NTA) (Figure 1B). Exosomes were further confirmed by the presence of protein markers such as CD63 and TSG101, and the absence of a negative marker GAPDH (Figure 1C). PKH67-labelled SMSCs-Exos fused to RA-FLS, as determined by fluorescence microscopy (Figure 1D). We then investigated the effects of SMSCs-Exos on RA-FLS-induced angiogenesis. To mimic the inflamed micro-environment of synovial tissue, IL-6 and its soluble receptor (sIL-6R) were added to the co-culture of RA-FLS and HDMECs, and the effects of SMSCs-Exos on RA-FLS-induced angiogenesis were investigated. After 48 h of incubation with IL-6+sIL-6R, VEGF mRNA (Figure 1E) and protein expression (Figure 1F) in co-cultured RA-FLS were significantly upregulated, and this upregulation was significantly inhibited by SMSCs-Exos. However, SMSCs-Exos did not significantly alter VEGF expression without IL-6+sIL-6R treatment. The expression of VEGF protein in the supernatants from the co-culture of RA-FLS and HDMECs showed a similar trend as those from the cell lysate of co-cultured RA-FLS (Figure 1G). Next, we asked whether VEGF secreted from human RA-FLS into the co-culture supernatants would promote the angiogenic activity of HDMECs. HDMECs were treated with supernatants from co-cultures, and the vascular structures formed by treated cells were carefully examined after exosomes were removed. HDMECs showed significant changes in migration by the Transwell assay (Figure 1H), the capillary-like structure formation by the tube formation test (Figure 1I), and microvessel sprouting by the ex vivo aortic ring angiogenesis assay (Figure 1J). These observations are consistent with the changes in VEGF expression in supernatants from the co-cultures of RA-FLS and HDMECs.
The Identification and Characteristics of circRNAs in SMSCs-Exos
Ten most upregulated circRNAs in MSCs compared with their differentiated stromal cell counterparts were reported previously.18 Using qRT–PCR, 4 circRNAs were validated to be expressed at distinctly higher levels in SMSCs compared to RA-FLS (Supplementary Figure 3A), and all 4 circRNAs, unlike linear RNA or actin, were resistant to digestion by RNase R (Supplementary Figure 3B). We further confirmed that hsa_circ_0073244 (circEDIL3) was the most highly expressed circRNA among the above four circRNAs in SMSCs-Exos (Supplementary Figure 4A). Characterization of hsa_circ_0073244 is shown in Supplementary Figure 4B. Thus, we suspected that circRNA could be transferred into RA-FLS as a cargo in SMSCs-Exos and might account for the biological functions of SMSCs-Exos.
To explore whether SMSCs-derived exosomal circRNA could affect VEGF secretion by synoviocytes, SMSCs stably overexpressing circEDIL3 (Ad-circEDIL3-SMSCs) were constructed by adenovirus infection, and SMSCs-Exos were extracted from the culture supernatants. Overexpression of circEDIL3 was confirmed by qRT–PCR (Supplementary Figure 4C). The exosomes were carefully extracted from the culture supernatants of Ad-circEDIL3-transfected SMSCs (Ad-circEDIL3-SMSCs-Exos). The expression of circEDIL3 was markedly elevated in Ad-circEDIL3-SMSCs-Exos (Supplementary Figure 4D). Ad-circEDIL3-SMSCs-Exos and its corresponding vector control (Ad-Vector-SMSCs-Exos) were added to the co-culture of RA-FLS and HDMECs with/without IL-6+sIL-6R. We found that Ad-circEDIL3-SMSCs-Exo treatment significantly reduced the levels of VEGF mRNA and protein in RA-FLS as well as VEGF concentration in the supernatant of the co-cultures compared with Ad-Vector-SMSCs-Exo treatment when the culture was induced with IL-6+sIL-6R; however, VEGF levels did not change significantly after Ad-circEDIL3-SMSCs-Exo treatment in the absence of IL-6+sIL-6R (Figure 2A-C). In summary, we examined circEDIL3 in SMSCs-Exos and proved that circEDIL3 was abundant and stable in SMSCs-Exos, and that this circular RNA might be transferred to RA-FLS by SMSCs-Exos to further inhibit VEGF production by RA-FLS in an IL-6-induced inflammatory environment.
Regulation of VEGF Expression by circEDIL3 via the miR-485-3p/PIAS3/STAT3 Axis in RA-FLS
Recent studies have shown that the main regulatory mechanism of circular RNA is to function as a miRNA sponge, and that this mainly occurs in the cytoplasm.7 Fluorescence in situ hybridization (FISH) results showed that most circEDIL3 transcript signals were located in the cytoplasm of SMSCs, with a few hybridization signals located in the nuclear zone (Supplementary Figure 5). As circEDIL3 has been confirmed to be localized in the cytoplasm, we wondered whether circEDIL3 would also act as a miRNA sponge. MicroRNAs “sponged” by circEDIL3 were searched using Arraystar Proprietary Algorithms based on the public databases TargetScan and miRanda, and the highest ranking candidate miRNAs were selected. At least one miRNA-binding site identified by the Arraystar Proprietary Algorithms was confirmed using the CircInteractome database (Supplementary Figure 6A). To validate whether the selected miRNAs could interact with circEDIL3, a luciferase reporter assay was performed. The sequence of circEDIL3 was inserted immediately downstream of the Renilla luciferase reporter gene, and each candidate interacting miRNA was co-transfected with the luciferase reporter into RA-FLS. MiR-485-3p, miR-198 and miR-326 reduced Renilla luciferase reporter activity compared to the negative control (Supplementary Figure 6B). Next, the levels of circEDIL3 were manipulated in RA-FLS and the efficiency of knockdown or overexpression was confirmed (Supplementary Figure 6C and D). qRT–PCR analysis showed the opposing trends in the expression of miR-485-3p and circEDIL3 (Figure 2D); other candidate miRNAs also had similar effects, but miR-485-3p showed the most pronounced effect (Supplementary Figure 6E). The interaction of circEDIL3 with its potential target miR-485-3p was further confirmed. The level of circEDIL3 expression was significantly reduced/enhanced by miR-485-3p mimics/inhibitor compared with respective controls (Figure 2E). Next, the miRNA target site in the circEDIL3 sequence was mutated (Supplementary Figure 7A), and the Renilla luciferase assay was repeated. No significant change in luciferase levels was observed, demonstrating the specificity of the interaction between circEDIL3 and miR-485-3p target site (Figure 2F). Using a pull-down assay with biotin-labelled oligonucleotides complementary to the back-splice junction of circEDIL3, the direct interaction between miR-485-3p and circEDIL3 in its native circular structure was validated. Consistent with the Renilla luciferase reporter assay, the circEDIL3-specific probe, but not the control probe, pulled down significant amount of miR-485-3p (Figure 2G). We also used a 3ʹ end biotin-labelled miR-485-3p to pull down circRNAs and confirmed a significant enrichment of circEDIL3 in the pulled-down products (Figure 2H). The bioinformatics database StarBase predicted that the 3ʹ UTR of human PIAS3 harboured a putative binding site of miR-485–3p (Supplementary Figure 6A). The luciferase plasmids pGL3-PIAS3-WT and pGL3-PIAS3-Mut were constructed (Supplementary Figure 7B) and co-transfected with miR-485-3p mimics into RA-FLS. Luciferase activity of the PIAS3 wild-type reporter was inhibited, but the activity of the mutated luciferase reporter remained unchanged (Figure 2I). Furthermore, we employed the biotinylated RNA pull-down assay to confirm the results. The PIAS3 mRNA captured by biotin-miR-485-3p was prominently increased compared with the control (Figure 2H). PIAS3 mRNA expression in RA-FLS was determined by qRT–PCR. As shown in Figure 3A and B, the expression of PIAS3 mRNA and protein was strongly inhibited/promoted by miR-485-3p mimics/inhibitor in RA-FLS. Together, the above results indicated that PIAS3 might be a potential target gene of miR-485-3p and that circEDIL3 could regulate PIAS3 by sponging miR-485-3p.
CircEDIL3 Regulates STAT3 Activity via the miR-485-3p/PIAS3 Axis
PIAS3 can bind to p-STAT3 to antagonize the function of STAT3, which is a key transcription factor of VEGF. Therefore, we speculated that circEDIL3 might regulate STAT3 activity via the miR-485-3p/PIAS3 axis. To verify this hypothesis, the protein level of PIAS3 and the p-STAT3/STAT3 ratio were examined by Western blot analysis upon miR-485-3p overexpression or knockdown. As shown in Figure 3B, PIAS3 protein expression significantly decreased when miR-485-3p mimics were added, while an opposite effect was observed on p-STAT3 expression; however, the protein level of STAT3 did not change significantly, indicating that STAT3 activity, but not the protein level, was elevated. Furthermore, the miR-485-3p inhibitor played an opposite role in PIAS3 protein expression and STAT3 activity.
Next, PIAS3 was overexpressed in the cells treated with miR-485-3p mimics. PIAS3 was successfully overexpressed at the mRNA and protein levels (Figure 3C and D) Despite suppression by miR-485-3p mimics, PIAS3 protein expression was upregulated by PIAS3 overexpression. The increase in p-STAT3 induced by miR-485-3p mimics was blocked by PIAS3 overexpression, and there was no significant change in the STAT3 protein level.
Finally, cells were subjected to a combination of miR-485-3p inhibitor treatment and circEDIL3 knockdown. Si-circEDIL3 inhibited the increase in PIAS3 protein expression as well as the decrease in p-STAT3 expression induced by the miR-485-3p inhibitor without a significant change in STAT3 protein expression (Figure 3F), indicating downregulation of circEDIL3 restored STAT3 activity.
PIAS3 and STAT3 mRNA expression levels were also determined by qRT–PCR. The results showed that PIAS3 protein expression changed in parallel with its mRNA expression; however, there were no significant changes in STAT3 mRNA levels regardless of administration of miR-485-3p mimics/inhibitor, PIAS3 overexpression, and circEDIL3 knockdown either singly or in combination (Figure 3A, C and E).
SMSCs-Exo-circEDIL3 Directly Regulates VEGF Expression via the miR-485-3p/PIAS3/STAT3 Axis
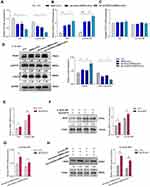
Ad-circEDIL3 or its corresponding negative control (Ad-Vector) was transfected into SMSCs, and exosomes were isolated from the culture media to deliver circEDIL3 (Ad-circEDIL3- SMSCs-Exos) or Vector (Ad-Vector-SMSCs-Exos). The results showed that regardless of IL-6+sIL-6R induction, Ad-circEDIL3-SMSCs-Exos suppressed the expression of miR-485-3p, increased PIAS3 mRNA, and the STAT3 mRNA level remained stable. Similar effects on miR-485-3p/PIAS3/STAT3 mRNA expression were observed in the SMSCs-Exos treated samples but not in PBS treated controls (Figure 4A-C).
Protein detection was performed by Western blot. Interestingly, regardless of IL-6+sIL-6R induction, the PIAS3 protein expression paralleled the PIAS3 mRNA level. Upon IL-6+sIL-6R induction, an opposing effects were observed on STAT3 activity and PIAS3 protein levels (Figure 4D). However, in the absence of IL-6+sIL-6R induction, the p-STAT3 protein level remained stable, and STAT3 activity, ie, the p-STAT3/STAT3 protein ratio did not change significantly while the level of PIAS3 protein significantly changed (Supplementary Figure 8).
To explore the actual role of STAT3 in the regulation of VEGF expression, STAT3 was transfected into RA-FLS using an adenovirus vector to augment intracellular STAT3. Under this condition, we found that p-STAT3 (Supplementary Figure 9) and VEGF mRNA and protein expression were significantly increased regardless of IL-6+sIL-6R induction (Figure 4E and F). Furthermore, Ad-STAT3 significantly rescued the downregulation of VEGF expression induced by SMSCs-Exos or Ad-circEDIL3-SMSCs-Exos under IL-6+sIL-6R induction (Figure 4G and H). These data further illustrated that circEDIL3 could be transferred across cells via SMSCs-Exos and then facilitate the expression of PIAS3 as a sponge of miR-485-3p, thereby upregulated STAT3 activity, accelerated VEGF expression and subsequent angiogenesis induced by RA-FLS.
Effect of SMSCs Exosomal circEDIL3 on Arthritis in CIA Mice
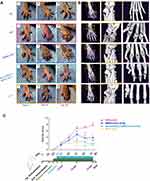
To further investigate the effect of exosomal circEDIL3 on arthritis in vivo, CIA mice were injected intraarticularly with Ad-circEDIL3-SMSCs-Exos, Ad-Vector-SMSCs-Exos or SMSCs-Exos. Exosome treatment lasted from Day 26 to Day 39 after the first (primary) immunization. We assessed the severity of arthritis from Day 24 after primary immunization according to a standard arthritic scoring system,4 and the scores of CIA mice were recorded every 3 d until the end of the experiment. Micro-CT images were taken and evaluated on Day 40. As shown in Figure 5A and B, CIA developed rapidly in paws after mice were immunized with Collagen II and clinical symptoms and joint destruction were evident. Methotrexate (MTX) treatment (1.5 mg/kg/week), as a positive control, had a strong inhibitory effect on the development of arthritis (Figure 5A-C). CIA mice treated with SMSCs-Exos continuously showed significantly reduced signs of arthritis, lower clinical arthritis scores and less joint destruction than CIA control mice (Figure 5A-C). Furthermore, hind paw thickness and clinical arthritis scores as well as bone erosion were significantly reduced in the CIA mice treated with Ad-circEDIL3-SMSCs-Exos relative to those in the SMSCs-Exos (Figure 5A-C) or Ad-Vector-SMSCs-Exo treatment groups (Supplementary Figure 10A and B). Representative clinical and micro-CT pictures of the paws from mice under different treatment groups are shown in Figure 5A and B. Evaluation of the arthritis score and the experimental schedule are shown in Figure 5C. Mean micro-CT values were evaluated and are shown in Supplementary Figure 10B. The mean body weight of the CIA control group decreased significantly compared to that of normal mice from Day 24 to Day 39. However, the mean body weights of the groups treated with Ad-circEDIL3-SMSCs-Exos, Ad-Vector-SMSCs-Exos and SMSCs-Exos were significantly more than those treated with PBS, and there was no significant difference compared to normal mice (Supplementary Figure 10C).
Histological Analysis
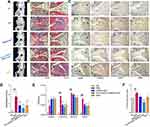
Knee joints from CIA mice on Day 40 were evaluated by micro-CT and histopathologic assessment of HE stained sections. The micro-CT results showed destruction of joint structure and rough bone surface in mice in the CIA group. Joint damage was significantly mitigated in the SMSCs-Exo group compared with the PBS-treated group. Notably, knee joints from the Ad-circEDIL3-SMSCs-Exo-injected mice showed a remarkable improvement in arthritis compared with the SMSCs-Exo and Ad-Vector-SMSCs-Exo groups (Figure 6A and Supplementary Figure 10D). HE staining analysis showed that after induction of CIA, mice exhibited massive inflammatory infiltration, synovial hyperplasia and pannus formation in the synovial tissue; leukocyte infiltration and destruction of bone and cartilage were significantly ameliorated in the SMSCs-Exo and Ad-circEDIL3-SMSCs-Exo groups compared with the CIA control group. The improvement of joint pathological changes was more evident in the Ad-circEDIL3-SMSCs-Exo group than in the SMSCs-Exo group (Figure 6B and D). Overall, joint damages demonstrated by micro-CT and histological manifestations were consistent with the severity of clinical symptoms and arthritis scores of CIA mice. Moreover, immunohistochemical staining of joint tissue sections and average integrated optical density (IOD) values showed the decreased staining intensity of PIAS3, increased ratio of p-STAT3/STAT3, and increased staining intensity of VEGF in CIA control mice compared with those in normal control mice. SMSCs-Exo treatment elevated the staining intensity of PIAS3 and VEGF and reduced the ratio of p-STAT3/STAT3 compared with those in CIA control mice. Moreover, immunohistochemical staining of PIAS3 was significantly stronger, the p-STAT3/STAT3 ratio reduced, and VEGF stronger in the Ad-circEDIL3-SMSCs-Exo group than in the mice treated with SMSCs-Exos. The trends of the changes of staining intensity of PIAS3/p-STAT3/STAT3/VEGF in the MTX-treated group were similar to those in mice treated with Ad-circEDIL3-SMSCs-Exos (Figure 6C, E and F).
Discussion
CircRNAs have been previously reported to be involved in the proinflammatory state in RA patients.19 However, the relationship between circRNAs and inflammation-induced angiogenesis has not been determined. STAT3 signalling has recently emerged as a main regulator of inflammation, and early suppression of STAT3 correlates well with longer-term clinical benefit in RA.20 In our study, one circRNA (hsa_circ_0073244, circEDIL3) was found to target the core molecule STAT3 and then inhibit downstream VEGF, thus playing a critical role in both inflammatory and angiogenic events. This contrast with other circRNAs previously reported because it does not simply directly target and promote VEGF molecules to induce angiogenesis.21,22 VEGF-induced angiogenesis in the inflammatory microenvironment remains a considerable challenge facing the treatment of RA and even numerous other diseases related to inflammation and angiogenesis.23,24 To our knowledge, our discovery of this key circRNA is the first be involved in both inflammation and angiogenesis. Therefore, this finding would provide a more specific therapeutic target and benefit precision therapy.
More interestingly, circEDIL3, one of the most highly expressed circRNAs in SMSCs, could be delivered into recipient RA-FLS via exosomes from SMSCs (SMSCs-Exos), which repressed VEGF expression in the cell lysate of co-cultured RA-FLS as well as in the co-culture supernatant of RA-FLS and HDMECs and reduced vascularity. Furthermore, a decrease in VEGF expression in co-cultured RA-FLS lysate was observed after the addition of circEDIL3-overexpressing SMSCs-Exos, suggesting the feasibility of a novel exosomal therapy targeting circRNA. This discovery of the therapeutic potential of SMSCs-Exos on the inhibition of abnormal angiogenesis was in accordance with a previous report;13,25 however, circRNAs involved in the antiangiogenic effect of SMSCs-Exos during inflammation have not yet been reported. Due to the stable cyclic structure,7 circRNA delivery will be more promising for treatment than that of other noncoding RNAs, and circRNA-overexpressing SMSCs-Exos, which might further protect the RNA cargo from degradation, are expected to be a new therapeutic method. This approach may have greater advantages over miRNA-expressing MSC-Exos that have been reported for therapy.13
To verify the interdependent regulatory interactions among circEDIL3 and other molecules in detail, we examined the expression levels after either overexpression or knockdown in vitro. We observed that the level of miR-485-3p was significantly increased/reduced after circEDIL3 knockdown/overexpression, and the level of circEDIL3 could be significantly reduced/enhanced by miR-485-3p mimics/inhibitor. The expression of PIAS3 was significantly decreased/increased by miR-485-3p mimics/inhibitor, respectively. The downregulation/upregulation of PIAS3 expression by miR-485-3p mimics/inhibitor was rescued by PIAS3 overexpression/circEDIL3 knockdown. Interestingly, the biological activity of STAT3, ie, the p-STAT3/STAT3 ratio showed opposing changes with PIAS3 levels, without discernable changes in STAT3 mRNA expression. These results indicated the existence of the circEDIL3/miR-485-3p/PIAS3/STAT3 functional module (Figure 7) and confirmed that PIAS3 was a cellular inhibitor of STAT3, consistent with earlier observations in different cell models.26,27 Although recent developments regarding the JAK/STAT3 signalling pathway as a therapeutic target, such as JAK2 inhibitors, have proven to be effective in the treatment of RA,28 many patients still have poor responses to treatment due to the heterogeneity of patients. In this study, we focused on PIAS3, an inhibitor of STAT3, and modulated this Rho GTPase family member instead of blocking the JAK pathway. Our study provides an alternative to currently available treatment options, and of course, it will be superior to the previous option. Since different people are more sensitive to different drugs, precision therapy will produce better results.
CircEDIL3 overexpression upregulated PIAS3 protein expression, which, in turn, inactivated STAT3 to alleviate RA disease progression. This is the first report on the circRNAs relevant to PIAS3 and thus STAT3 inhibition in arthritis therapy. The benefit of increased PIAS3 expression on RA was consistent with the results of Samarpita et al29,30 but different from those of Lao et al31 possibly due to the different status of STAT3 activity due to distinct disease activity or different tissue contexts. Furthermore, we found that the expression of VEGF may be elevated by STAT3 overexpression, suggesting that VEGF is the downstream mediator of the effects of STAT3, in accordance with a previous study.32,33 As STAT3 is a key transcription factor in inflammation and other pathological events, such as angiogenesis, in various diseases,34,35 discovery of the circEDIL3/miR-485-3p/PIAS3/STAT3 functional module may also provide a novel target for the treatment of other diseases in which STAT3 plays a central role.
Surprisingly, in this study, we also found that, without inflammatory induction by the IL-6/sIL-6R complex, the expression of VEGF in the cell lysate of co-cultured RA-FLS and the supernatant from the co-culture of RA-FLS and HDMECs showed no significant change with SMSCs-Exo treatment, although levels of circEDIL3, miR-485-3p, and PIAS3 changed consistently after SMSCs-Exo treatment regardless of IL-6+sIL-6R addition. Interestingly, STAT3 activity was significantly upregulated after IL-6+sIL-6R induction, in agreement with a previous report,36 and in the absence of IL-6+sIL-6R induction, STAT3 activity was not significantly altered, although circEDIL3, miR-485-3p, and PIAS3 were already altered. The above results indicated that IL-6 induction was a significant promoter of STAT3 activity, and the direct inhibitory effect of PIAS3 on STAT3 was dependent on the activity of STAT3. Perhaps this might provide an explanation of why SMSCs-Exo treatment stimulated VEGF-induced angiogenesis in the regenerated tissue37, where a marked increase in IL-6 expression was absent. Therefore, the effect of SMSCs-Exos on angogenesis may closely depend on STAT3 activity in different tissue contexts. The findings of this study will undoubtedly make our understanding of the effect of SMSCs-Exos on angiogenesis more comprehensive and will be of great help for the rational application of SMSCs-Exos in the future treatment.
Another significance our present study is that we demonstrated the the clinical potential of SMSCs-Exos produced by bioengineering as a drug carrier38 which can “kill two birds with one stone”. In this study, SMSCs-Exos had the advantages of exerting anti-inflammatory effects and blocking their associated angiogenesis. The positive effect of targeting STAT3 might make them more suitable for the treatment of RA or other diseases, such as cancer, as carriers of STAT3 antagonists including JAK inhibitors, miRNAs targeting STAT3 mRNA,39 and other chemical drugs including arsenic trioxide4,17,40–42, with similar antiangiogenic effects. SMSCs-Exos might function synergistically with the loaded drugs, thus decreasing drug resistance and reducing the dosage of the drug cargo, achieving equivalent or even superior therapeutic effects, and significantly reducing the toxic side effects of the loaded drugs. We will continue to research the optimal dose ratio of SMSCs-Exos to loaded drugs, and further steps will be taken to study how to increase the yields of exosomes for mass production.
In conclusion, circEDIL3 derived from SMSCs-Exos was found to suppress inflammation-induced angiogenesis, which promotes pannus progression, through the miR-485-3p/PIAS3/STAT3/VEGF functional module both in vitro and in vivo, thus ameliorated RA (Figure 7). Moreover, circEDIL3-overexpressing SMSCs-Exos might serve as valuable agents for the development of novel precision therapeutic strategies for RA, either as monotherapy or in combination treatment as novel drug carriers.
Abbreviations
RA, rheumatoid arthritis; circRNAs, circular RNAs; miRNAs, microRNAs; MSCs, mesenchymal stem cells; SMSCs, synovial mesenchymal stem cells; SMSCs-Exos, SMSCs derived exsomes; Ad-circEDIL3-SMSCs, circEDIL3 expression adenovirus transfected SMSCs; Ad-circEDIL3-SMSCs-Exos, exosomes secreted from Ad-circEDIL3-SMSCs; RA-FLS, fibroblast-like synoviocytes from RA patients; HDMECs, human dermal microvascular endothelial cells; CII, bovine type II collagen; CIA, collagen-induced arthritis; VEGF, vascular endothelial growth factor; STAT3, signal transducer and activator of transcription 3; PIAS3, protein inhibitor of activated STAT3; TEM, transmission electron microscope; NTA, nanoparticle tracking analysis; sIL-6R, IL-6 and its soluble receptor; FISH, fluorescence in situ hybridization; MTX, methotrexate; IOD, integrated optical density; FCM, flow cytometry; CFA, complete Freund’s adjuvant; micro-CT, micro-computed tomography; H&E, hematoxylin-eosin.
Ethical Approval Information
Ethics Committee of the First Affiliated Hospital of Harbin Medical University.
Acknowledgments
This work is supported by National Natural Science Foundation of China (NSFC 81901638), Chinese Postdoctoral Science Foundation (Grant No. 2019M651309), Heilongjiang Provincial Postdoctoral Science Foundation (Grant No. LBH-Z18226) and Scientific Research Innovation Foundation of the First Affiliated Hospital of Harbin Medical University (Grant No. 2019B18) to Juan Zhang, and Young and Middle-aged People Scientific Research Innovation Foundation of the Second Affiliated Hospital of Harbin Medical University (Grant No. KYCX2019-10) to Maolin Chu.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kurosaka D, Yoshida K, Yasuda J, et al. Inhibition of arthritis by systemic administration of endostatin in passive murine collagen induced arthritis. Ann Rheum Dis. 2003;62(7):677–679. doi:10.1136/ard.62.7.677
2. Wang W, Li C, Zhang Z, Zhang Y. Arsenic trioxide in synergy with Vitamin D rescues the defective VDR-PPAR-γ functional module of autophagy in rheumatoid arthritis. PPAR Res. 2019;2019:6403504.
3. Li C, Zhang J, Wang W, Wang H, Zhang Y, Zhang Z. Arsenic trioxide improves Treg and Th17 balance by modulating STAT3 in treatment-naïve rheumatoid arthritis patients. Int Immunopharmacol. 2019;73:539–551.
4. Zhang J, Li C, Zheng Y, Lin Z, Zhang Y, Zhang Z. Inhibition of angiogenesis by arsenic trioxide via TSP-1-TGF-β1-CTGF-VEGF functional module in rheumatoid arthritis. Oncotarget. 2017;8(43):73529–73546.
5. Ilson DH. Advances in the treatment of gastric cancer. Curr Opin Gastroenterol. 2017;33(6):473–476.
6. Liu CX, Li X, Nan F, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell. 2019;177(4):865–880.e821.
7. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
8. Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21(1):22–36.
9. Kreuger J, Phillipson M. Targeting vascular and leukocyte communication in angiogenesis, inflammation and fibrosis. Nat Rev Drug Discov. 2016;15(2):125–142.
10. Xie Y, Dang W, Zhang S, et al. The role of exosomal noncoding RNAs in cancer. Mol Cancer. 2019;18(1):37.
11. Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116.
12. Shahir M, Mahmoud Hashemi S, Asadirad A, et al. Effect of mesenchymal stem cell-derived exosomes on the induction of mouse tolerogenic dendritic cells. J Cell Physiol. 2020;235(10):7043–7055.
13. Chen Z, Wang H, Xia Y, Yan F, Lu Y. Therapeutic potential of mesenchymal cell-derived miRNA-150-5p-expressing exosomes in rheumatoid arthritis mediated by the modulation of MMP14 and VEGF. J Immunol. 2018;201(8):2472–2482.
14. De Bari C, Dell’Accio F, Tylzanowski P, Luyten FP. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthr Rheum. 2001;44(8):1928–1942.
15. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659.
16. Yang X, Lin A, Jiang N, et al. Interleukin-6 trans-signalling induces vascular endothelial growth factor synthesis partly via Janus kinases-STAT3 pathway in human mesothelial cells. Nephrology. 2017;22(2):150–158.
17. Chu M, Zhang C. Inhibition of angiogenesis by leflunomide via targeting the soluble ephrin-A1/EphA2 system in bladder cancer. Sci Rep. 2018;8(1):1539.
18. Cherubini A, Barilani M, Rossi RL, et al. FOXP1 circular RNA sustains mesenchymal stem cell identity via microRNA inhibition. Nucl Acids Res. 2019;47(10):5325–5340.
19. Yang J, Cheng M, Gu B, Wang J, Yan S, Xu D. CircRNA_09505 aggravates inflammation and joint damage in collagen-induced arthritis mice via miR-6089/AKT1/NF-κB axis. Cell Death Dis. 2020;11(10):833.
20. Boyle DL, Soma K, Hodge J, et al. The JAK inhibitor tofacitinib suppresses synovial JAK1-STAT signalling in rheumatoid arthritis. Ann Rheum Dis. 2015;74(6):1311–1316.
21. Zou J, Liu KC, Wang WP, Xu Y. Circular RNA COL1A2 promotes angiogenesis via regulating miR-29b/VEGF axis in diabetic retinopathy. Life Sci. 2020;256:117888.
22. Guo J, Chen M, Ai G, Mao W, Li H, Zhou J. Hsa_circ_0023404 enhances cervical cancer metastasis and chemoresistance through VEGFA and autophagy signaling by sponging miR-5047. Biomed Pharmacother/Biomedecine & Pharmacotherapie. 2019;115:108957.
23. Zhang J, Chu M. Differential roles of VEGF: relevance to tissue fibrosis. J Cell Biochem. 2019;120(7):10945.
24. Saharinen P, Eklund L, Alitalo K. Therapeutic targeting of the angiopoietin-TIE pathway. Nat Rev Drug Discov. 2017;16(9):635–661.
25. Pakravan K, Babashah S, Sadeghizadeh M, et al. MicroRNA-100 shuttled by mesenchymal stem cell-derived exosomes suppresses in vitro angiogenesis through modulating the mTOR/HIF-1α/VEGF signaling axis in breast cancer cells. Cell Oncol. 2017;40(5):457–470.
26. Jiao J, Zhang R, Li Z, et al. Nuclear Smad6 promotes gliomagenesis by negatively regulating PIAS3-mediated STAT3 inhibition. Nat Communi. 2018;9(1):2504.
27. Polimeno L, Francavilla A, Piscitelli D, et al. The role of PIAS3, p-STAT3 and ALR in colorectal cancer: new translational molecular features for an old disease. Eur Rev Med Pharmacolo Sci. 2020;24(20):10496–10511.
28. Malemud CJ. The role of the JAK/STAT signal pathway in rheumatoid arthritis. Ther Advan Musculoskel Dis. 2018;10(5–6):117–127.
29. Samarpita S, Ganesan R, Rasool M. Cyanidin prevents the hyperproliferative potential of fibroblast-like synoviocytes and disease progression via targeting IL-17A cytokine signalling in rheumatoid arthritis. Toxicol Appl Pharmacol. 2020;391:114917.
30. Tang X, Yin K, Zhu H, et al. Correlation between the expression of MicroRNA-301a-3p and the proportion of Th17 cells in patients with rheumatoid arthritis. Inflammation. 2016;39(2):759–767.
31. Lao M, Shi M, Zou Y, et al. Protein inhibitor of activated STAT3 regulates migration, invasion, and activation of fibroblast-like synoviocytes in rheumatoid arthritis. J Immunol. 2016;196(2):596–606.
32. Chen X, Zheng Q, Li W, et al. SOX5 induces lung adenocarcinoma angiogenesis by inducing the expression of VEGF through STAT3 signaling. OncoTargets Ther. 2018;11:5733–5741.
33. Chou JC, Lieu FK, Ho DM, et al. Regulation of extracellular and intracellular prolactin on cell proliferation and survival rate through GHR/JAK2/STAT3 pathway in NSCLC. Chemosphere. 2021;264(Pt 1):128604.
34. Hu YS, Han X, Liu XH. STAT3: a potential drug target for tumor and inflammation. Curr Top Med Chem. 2019;19(15):1305–1317.
35. Chen Q, Lv J, Yang W, et al. Targeted inhibition of STAT3 as a potential treatment strategy for atherosclerosis. Theranostics. 2019;9(22):6424–6442.
36. Hirano T. IL-6 in inflammation, autoimmunity and cancer. Int Immunol. 2021;33(3):127–148.
37. Han Y, Ren J, Bai Y, Pei X, Han Y. Exosomes from hypoxia-treated human adipose-derived mesenchymal stem cells enhance angiogenesis through VEGF/VEGF-R. Int J Biochem Cell Biol. 2019;109:59–68.
38. Lin Y, Wu J, Gu W, et al. Exosome-Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Advan Sci. 2018;5(4):1700611.
39. Lu FB, Chen DZ, Chen L, et al. Attenuation of experimental autoimmune hepatitis in mice with bone mesenchymal stem cell-derived exosomes carrying MicroRNA-223-3p. Mol Cells. 2019;42(12):906–918.
40. Zhang J, Zhang Y, Wang W, Zhang Z. Potential molecular mechanisms underlying the effect of arsenic on angiogenesis. Arch Pharm Res. 2019;42(11):962–976.
41. Zhang J, Zhang Y, Wang W, Li C, Zhang Z. Double-sided personality: effects of arsenic trioxide on inflammation. Inflammation. 2018;41(4):1128–1134.
42. Zhang C, Chu M. Leflunomide: a promising drug with good antitumor potential. Biochem Biophys Res Commun. 2018;496(2):726–730.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.