Back to Journals » International Journal of General Medicine » Volume 15
The Value of Serum miR-139-3p Expression Level in Predicting Postoperative Survival of Colon Cancer Patients
Received 6 November 2021
Accepted for publication 19 January 2022
Published 11 February 2022 Volume 2022:15 Pages 1405—1412
DOI https://doi.org/10.2147/IJGM.S346674
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Guohong Fu,1,* Haimei Xu,2,* Cong Zhou3
1Department of General Surgery, Sanya Central Hospital, Sanya, Hainan, People’s Republic of China; 2Department of Gastroenterology, People’s Hospital of Danzhou, Danzhou, Hainan, People’s Republic of China; 3Department of General Surgery, People’s Hospital of Danzhou, Danzhou, Hainan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Cong Zhou, Department of General Surgery, Danzhou People’s Hospital, 21-1 Datong Road, Danzhou, 571179, People’s Republic of China, Tel/Fax +8615851928641, Email [email protected]
Background: To explore the value of serum miR-139-3p expression level in predicting postoperative survival of colon cancer patients.
Methods: We selected 158 cases enrolled in our hospital from January 2017 to December 2019. Using real-time fluorescence quantitative PCR, the expression extents of serum miR-139-3p among patients suffering from colon cancer were detected. The enrollment of patients was performed in the high or low miR-139-3p group on the basis of the cutoff value determined by ROC curve analysis. The risk elements influencing the postoperative survival of colon cancer patients were analyzed by the Kaplan–Meier approach and univariate and multivariate Cox regression models.
Results: Compared with control group, significantly lower expression level of serum miR-139-3p was shown in colon cancer group (P < 0.05). Its low expression of miR-139-3p was associated with TNM stage, degree of differentiation, tumor sizes, lymph node metastasis and vascular infiltration in patients with colon cancer (all P < 0.05), which was also significantly associated with short survival time of colon cancer patients (P < 0.05). Multivariate Cox regression model analysis displayed that TNM phase, lymph node metastasis and miR-139-3p < 2.17 were independent risk elements affecting postoperative survival (P < 0.05).
Conclusion: The low expression level of miR-139-3p is related to the short survival time of colon cancer patients, and it is expected to be used as a biological indicator to predict the postoperative survival of colon cancer.
Keywords: colon cancer, miR-139-3p, prognostic evaluation
Introduction
Colon cancer is the most common malignant tumor of the digestive system, causing about 700,000 deaths each year, accounting for about 10% of all cancers diagnosed globally.1,2 However, because the primary site of tumor is not clear, the patient’s symptoms are not obvious, and the prognosis of colon cancer is still unsatisfactory. Since early diagnosis of colon cancer is difficult, finding a reliable biomarker is crucial for the early diagnosis and prognostic evaluation. In recent years, a large number of studies have shown that microRNA (miRNA) plays a predominant role in the occurrence and development of cancer, and it participates in various cell physiological activities.3–6 miRNA is also considered as a candidate biomarker for cancer. Recent studies have found that miR-139-3p is abnormally expressed in gastrointestinal tumors and is involved in the pathogenesis of gastrointestinal tumors.7,8 There are few reports on the relationship between the expression level of miR-139-3p and the prognosis of colon cancer. In this study, the serum miR-139-3p expression level in patients with colon cancer was detected, and the relationship between its expression and postoperative survival of colon cancer patients was analyzed, in order to provide a reference for evaluating the prognosis of colon cancer and targeted therapy.
Materials and Methods
Research Objects
A total of 158 colon cancer patients admitted to our hospital from January 2017 to December 2019 were selected, including 87 males and 71 females, aged 31–77 (55.62±8.73) years old. Inclusion criteria: 1) Those who were diagnosed with colon cancer by pathological examination and all underwent elective radical resection of colon cancer; 2) Those who have not received anti-cancer treatment before operation. Exclusion criteria: 1) Those who cannot complete radical resection; 2) Patients with other malignant tumors. One hundred and fifty-eight cases of colon cancer were classified into 52 cases from stage I to II, 106 cases from stage III according to TNM stage; there were 55 cases of well-differentiated cancer and 103 cases of poorly differentiated cancer. Another 60 cases undergoing physical examination were enrolled as controls including 37 males and 23 females, with an average of (48.73±8.95) years. This study used TNM pathological staging, which is the most commonly used clinical staging method.
Research Methods
We use prospective research methods to follow up on 164 patients with colon cancer. The start time of follow-up is the first day after the radical resection of colon cancer. The patients are investigated by telephone or outpatient service, and the deadline is December 31, 2020. Record the overall survival (OS) and progression-free survival (PFS) of the patients. In this study, chemiluminescence assay was used to determine CEA level using Roche electrochemiluminescence immunoanalyzer and supporting kit. Positive criteria: CEA > 5 ng/mL was positive.
miR-139-3p Detection
MiR-139-3p primer design: Upstream: 5′-GCTAATCTGATGACTAGTCG-3′, downstream: 5′-AGCATTCATGCGTATGCG-3′. All patients took 5 mL of fasting venous blood and performed real-time fluorescent quantitative polymerase chain reaction (RT-PCR) on the ABI 7500 fluorescent quantitative PCR instrument (ABI Company, USA). Using U6 as internal reference. The systems and conditions for both reverse transcription reaction and amplification reaction were based on previous literature reported before.9
Statistical Methods
The statistical analyses were conducted through SPSS20.0 software. x±S was employed to represent the measurement data exhibiting a normal distribution. Comparisons between groups were analyzed with the independent sample T-test. Enumeration data between groups was compared with the χ2 test. Single-factor survival analysis and multivariate survival analysis were conducted by employing Kaplan-Meier, log-rank and Cox regression analyses.
Ethical Approval/Patient Consent
The present study was approved by the Ethics Committee of Danzhou People’s Hospital (number; 011/2016, dated 09/02/2016). Patients with colon cancer and healthy controls have approved the informed consent. This investigation was conducted based on the declaration of Helsinki.
Results
Expression Level and Best Cutoff Value
The expression level of serum miR-139-3p in the colon cancer group (1.94±0.63) was significantly lower than that of the control group (4.96±1.42), and the difference was statistically significant (t = 10.635, P < 0.01) (Figure 1). When miR-139-3p is 2.17, the Youden’s index is the largest: Taking 2.17 as the best cut-off point of miR-139-3p, its sensitivity is 87.5%, specificity is 80.6%, and the area under the curve is 0.871 (Figure 2).
 |
Figure 1 Comparison of serum levels of miR-139-3p between colon cancer group and control group. |
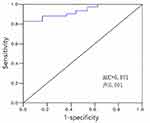 |
Figure 2 ROC curve of miR-139-3p for predicting survival of patients with colon cancer after operation. |
Clinicopathological Characteristics of Patients
Taking miR-139-3p=2.17 as the cutoff value, 158 patients with colon cancer were divided into high miR-139-3p group (miR-139-3p≥2.17, n = 65), low miR-139-3p group (miR −139-3p <2.17, n = 93). Compared the TNM stage, degree of differentiation, tumor size, lymph node metastasis and vascular invasion between high miR-139-3p group and low miR-139-3p group, the difference was statistically significant (P < 0.05); While there was no statistically significant difference in gender, age, pathological type and primary site between the two groups (P > 0.05) (Table 1).
 |
Table 1 Comparison of Clinical Features Between the High miR-139-3p Group and the Low miR-139-3p Group |
The Association Between the Expression Level and the Postoperative Survival
Of the 158 colon cancer patients, 5 were lost to follow-up, with a follow-up rate of 96.8% (153/158), the follow-up time was 9 to 67 months, and the median follow-up time was 29 months. Survival analysis showed that the overall survival rate of patients in the low miR-139-3p group was significantly lower than that in the high miR-139-3p group, the difference was statistically significant (50.4% vs 72.6%; Log-rank χ2=9.102, P < 0.01) (Figure 3); The progression-free survival rate of patients in the low miR-139-3p group was significantly lower than that in the high miR-139-3p group, and the difference was statistically significant (28.4% vs 59.7%; Log-rank χ2=11.705, P < 0.01) (Figure 4). We import the statistically significant factors in the univariate analysis into the COX regression analysis model. The results showed that TNM stage, lymph node metastasis, CEA (+) and miR-139-3p <2.17 were independent risk factors affecting the postoperative survival of colon cancer patients [HR (95% CI) = 2.683 (1.827~3.986), 2.914 (2.106~4.715), 3.916 (3.112~9.127)] (Table 2).
 |
Table 2 Univariate and Multivariate Cox Regression Analysis of Patients with Colon Cancer After Surgery |
 |
Figure 3 Total survival time curves of colon cancer patients in high miR-139-3p group and low miR-139-3p group. |
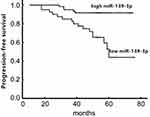 |
Figure 4 Comparison of progression-free survival curves between high and low miR-139-3p colon cancer patients. |
Discussion
Colon cancer is the most common gastrointestinal malignancy. Its incidence rate is second only to lung cancer and breast cancer. Recently, the incidence rate and mortality rate of colon cancer have been increasing speedily, which severely endangers human health and life. The occurrence and growth of colon cancer is a complex process, which is correlated to many factors, including polyps, inflammatory bowel disease, high-fat diet, shortage of exercise, drinking, smoking and obesity.10 Nonetheless, about half of colon cancer patients do not have polyp formation, inflammation or intestinal diseases, and not all people with bad habits such as high-fat diet, shortage of exercise, drinking, smoking and obesity will suffer from colon cancer. These findings indicate that other genetic or non-genetic elements regulating the risk of colon cancer have not been found. Radical treatment surgery is one of the effective means to cure colon cancer. However, postoperative local recurrence and metastasis rate is as high as 39%~75%, which seriously affects the treatment effect of colon cancer patients.11,12 The sensitivity and specificity of traditional tumor markers for colon cancer are not high. Hence, it is greatly significant to find a new marker to accurately predict the postoperative survival of colon cancer patients.
miRNA is a cluster of about 21–25 nucleotides that regulates gene expression by targeting mRNA. miRNA is in small volume, conserved, non-coding and endogenous.13,14 miRNA takes part in many biological and pathological processes consisting of cell development, signal transduction, cell propagation, migration and apoptosis.15–17 Abnormal expression of miRNAs is common. According to a large number of researches, miRNA expression is abnormal in body fluids of colon cancer patients including serum, plasma and feces.16–18 miRNA has been considered as a diagnostic and prognostic biomarker for the evaluation of the appearance, growth and prognosis of colon cancer. Various miRNAs can inhibit and promote cancer. In addition, miRNA exerts an important effect on colon cancer-related signal pathways including Wnt/b-catenin, EGFR, TGF-b, TP53.18 Researchers have shown that miRNA is abnormally expressed in colon cancer and closely associated with the appearance, growth, metastasis and prognosis of colon cancer.7,8 Recent research has confirmed that it exerts a great impact on the occurrence, invasion, metastasis, drug resistance and other pathological processes of malignant tumors by regulating a variety of signal pathways.10 Ng et al observed that the expression extent of colorectal cancer was noticeably decreased. It has a higher area under the curve, sensitivity and specificity for diagnosing colorectal cancer, and can be adopted as a noninvasive biomarker for diagnosing colorectal cancer.13
This study demonstrated that miR-139-3p may be a tumor suppressor gene, and its decreased expression may take part in the appearance and growth of colon cancer. The miR-139-3p may exert an anti-cancer effect on colon cancer. The studies of Feng et al displayed that miRNA in colon cancer tissues showed markedly lower expression than that in paired nearby non-cancer tissues and was closely linked to tumorigenesis and progression.14 It is a promising indicator for forecasting the prognosis of colon cancer. The studies of Zhang et al found that miRNA expression level was greatly related to TNM phase and lymph node metastasis, which could be employed as an independent factor to forecast the progression and prognosis of colon cancer.15 Other studies showed that miRNA combined with its related target genes regulated the proliferation, apoptosis and aggression of colon cancer cells, consequently conducting a significant effect on the clinical stage and metastasis of colon cancer.16 This study further explored the clinical value of miR-139-3p and the prognosis of patients suffering from colon cancer, and analyzed the relationships between the expression level of miR-139-3p and survival time. The results demonstrated that patients in low miR-139-3p group showed noticeably lower total survival rate than that in high miR-139-3p group, and that the disease-free survival rate of patients in low miR-139-3p group was dramatically lower than that in high miR-139-3p group, which indicated that the low expression of miR-139-3p was related to the short survival time of patients with colon cancer. Besides, multivariate Cox regression model analysis displayed that high TNM stage, lymph node metastasis and miR-139-3p <2.17 were independent risk elements influencing postoperative survival of colon cancer patients. The studies of Liu et al displayed that the level of miR-139-3p in colon cancer tissues was noticeably lower than that in nearby non-cancer tissues.17 A lower level of miR-139-3p is remarkably related to low overall survival rate. The decreased expression level of miR-139-3p can be employed as a biomarker to forecast the poor prognosis of colon cancer patients. Li et al showed that low expression of miRNA was related to poor prognosis of colon cancer patients.18 Compared with high expression of miRNA, low expression of miRNA had 1.92 times higher risk of short-term overall survival, suggesting that miRNA may be a potential biomarker to forecast the prognosis of colon cancer. The studies of Yan et al found that the low expression of miRNA had a significant impact on the total survival rate and progression-free survival rate of patients with colon cancer, which is expected to be a hopeful biomarker for the prognosis of colon cancer.19 Some researchers have also displayed that the abnormal expression of miRNA is greatly related to the clinical characteristics and survival rate of patients, and can be adopted to predict the survival rate and disease progression of patients with colon cancer.20
Although miR-139-3p is a hidden biomarker for the early diagnosis and prognosis of colon cancer patients, miR-139-3p still calls for further research. First, real-time fluorescence quantitative PCR detected the expression level of miR-139-3p in tumor cell lines and epithelial cell lines. Secondly, lentivirus vector was established by gene recombination technology, and lentivirus vector was transferred into cancer cell lines to produce a tumor cell line with decreased expression of miR-139-3p cell line.
Conclusion
In summary, the low expression level of miR-139-3p is related to the short survival time of colon cancer patients, and it is expected to be used as a biological indicator to predict the postoperative survival of colon cancer. In the future, more large-scale multi-center pre-treatment is urgently entailed forward-looking research to confirm our findings.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethical Statement
The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Acknowledgments
We sincerely thank some students from People’s Hospital of Danzhou for helping us in data collection and collation. Guohong Fu and Haimei Xu are co-first authors for this study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
The authors declare that there are no sources of funding to be acknowledged.
Disclosure
The authors declare that they have no competing interests.
References
1. Daly MC, Paquette IM. Surveillance, Epidemiology, and End Results (SEER) and SEER-medicare databases: use in clinical research for improving colorectal cancer outcomes. Clin Colon Rectal Surg. 2019;32(01):61–68. doi:10.1055/s-0038-1673355
2. Coebergh van den Braak RRJ, Sieuwerts AM, Lalmahomed ZS, et al. Confirmation of a metastasis-specific microRNA signature in primary colon cancer. Sci Rep. 2018;8:5242. doi:10.1038/s41598-018-22532-1
3. Chen J, Hu B, Wang W, et al. A six-microRNA signature to predict outcomes of patients with gastric cancer. FEBS Open Bio. 2019;9:538–547. doi:10.1002/2211-5463.12593
4. Chen WC, Lin MS, Ye YL, et al. microRNA expression pattern and its alteration following celecoxib intervention in human colorectal cancer. Exp Ther Med. 2012;3:1039–1048.
5. Ren Y, Shi G, Jiang P, Meng Q. MicroRNA-761 is downregulated in colorectal cancer and regulates tumor progression by targeting Rab3D. Exp Ther Med. 2019;17:1841–1846.
6. Zhu XL, Ren LF, Wang HP, et al. Plasma microRNAs as potential new biomarkers for early detection of early gastric cancer. World J Gastroenterol. 2019;25:1580–1591. doi:10.3748/wjg.v25.i13.1580
7. Chen F, Li Z, Zhou H. Identification of prognostic miRNA biomarkers for predicting overall survival of colon adenocarcinoma and bioinformatics analysis: a study based on the cancer genome atlas database. J Cell Biochem. 2019;120(6):9839–9849. doi:10.1002/jcb.28264
8. Xie B, Zhao R, Bai B, et al. Identification of key tumorigenesis-related genes and their microRNAs in colon cancer. Oncol Rep. 2018;40:3551–3560.
9. Chen X, Su X, Lin M, et al. Expression of miR-192-5p in colon cancer serum and its relationship with clinicopathologic features. Am J Transl Res. 2021;13:9371–9376.
10. Zhu Y, Zhou C, He Q. High miR-139-3p expression predicts a better prognosis for hepatocellular carcinoma: a pooled analysis. J Int Med Res. 2019;47(1):383–390. doi:10.1177/0300060518802727
11. Dougaz W, Bouasker I, Gouta EL, et al. Predictive factor of recurrence after curative resection for stage I–II colon cancer. Tunis Med. 2019;97:685–691.
12. Asano H, Kojima K, Ogino N, et al. Postoperative recurrence and risk factors of colorectal cancer perforation. Int J Colorectal Dis. 2017;32(3):419–424. doi:10.1007/s00384-016-2694-3
13. Ng L, Wan TM, Man JH, et al. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8:27393–27400. doi:10.18632/oncotarget.16171
14. Feng Y, Dong YW, Song YN, et al. MicroRNA449a is a potential predictor of colitis-associated colorectal cancer progression. Oncol Rep. 2018;40:1684–1694.
15. Zhang H, Wang Z, Ma R, Wu J, Feng J. MicroRNAs as biomarkers for the progression and prognosis of colon carcinoma. Int J Mol Med. 2018;42:2080–2088.
16. Jacob H, Stanisavljevic L, Storli KE, et al. Identification of a sixteen-microRNA signature as prognostic biomarker for stage II and III colon cancer. Oncotarget. 2017;8(50):87837–87847. doi:10.18632/oncotarget.21237
17. Liu X, Duan B, Dong Y, et al. MicroRNA-139-3p indicates a poor prognosis of colon cancer. Int J Clin Exp Pathol. 2014;7:8046–8052.
18. Li C, Yan G, Yin L, et al. Prognostic roles of microRNA 143 and microRNA 145 in colorectal cancer: a meta-analysis. Int J Biol Markers. 2019;34(1):6–14. doi:10.1177/1724600818807492
19. Yan S, Dang G, Zhang X, et al. Downregulation of circulating exosomal miR-638 predicts poor prognosis in colon cancer patients. Oncotarget. 2017;8(42):72220–72226. doi:10.18632/oncotarget.19689
20. Xu M, Kuang Y, Wang M, Han X, Yang Q. A microRNA expression signature as a predictor of survival for colon adenocarcinoma. Neoplasma. 2017;64(01):56–64. doi:10.4149/neo_2017_107
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
