Back to Journals » OncoTargets and Therapy » Volume 14
The Value and Clinical Significance of ZNF582 Gene Methylation in the Diagnosis of Cervical Cancer
Authors Zhang C, Fu S, Wang L, Wang F, Wu D, Zhe X, Xin H, Li H, Li D , Jin F, Shao R, Pan Z
Received 22 August 2020
Accepted for publication 18 November 2020
Published 14 January 2021 Volume 2021:14 Pages 403—411
DOI https://doi.org/10.2147/OTT.S277445
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Sanjeev K. Srivastava
Chunhe Zhang,1,* Shaowei Fu,1,* Luyue Wang,1,* Fang Wang,1 Dan Wu,1 Xiangyi Zhe,1 Huizhen Xin,1 Hongtao Li,1 Dongmei Li,1 Fuyuan Jin,1 Renfu Shao,2 Zemin Pan1
1Department of Biochemistry and Molecular Biology, School of Medicine, Shihezi University, Xinjiang Endemic and Ethnic Disease and Education Ministry Key Laboratory, Shihezi, Xinjiang 832002, People’s Republic of China; 2School of Science, Technology and Engineering, Genecology Research Centre, University of the Sunshine Coast, Sippy Downs, Queensland 4556, Australia
*These authors contributed equally to this work
Correspondence: Zemin Pan; Xiangyi Zhe
Department of Biochemistry and Molecular Biology, School of Medicine, Shihezi University, Xinjiang Endemic and Ethnic Disease and Education Ministry Key Laboratory, Shihezi, People’s Republic of China
Email [email protected]; [email protected]
Introduction: The aim of this study was to determine whether ZNF582 gene methylation and tissue protein expression can be used as a tool with high sensitivity and specificity for cervical cancer screening. We analyzed the correlation between promoter methylation of ZNF582 gene and cervical cancer and high risk HPV16/18 infection.
Methods: Tissue samples of normal cervical or chronic cervicitis (n=51), CIN (cervical intraepithelial neoplasia) (n=35), and cervical carcinoma (n=68) were tested for HPV16/18 infection by polymerase chain reaction (PCR). We also detected the methylation status of the ZNF582 gene promoter in the same tissues by methylation-specific PCR (MSP), then analyzed the correlation between ZNF582 promoter methylation and HPV16/18 infection. Immunohistochemistry was used to analyze ZNF582 gene expression in 152 cervical tissues. We detected ZNF582 mRNA expression in cervical tissues (including cancer and non-cancer) by real-time fluorescence quantitative PCR (qPCR).
Results: Among 93 high-grade cervical lesions (CINII and above) and cervical cancer samples, 57 cases were positive for HPV16/18 infection and 36 cases were negative. ZNF582 gene methylation occurred in 9 out of 51 cases in normal cervical tissues (17.6%), 16 of 35 cases in CIN tissues (45.7%), and 50 of 68 cases in cervical cancer (73.5%). The differences in methylation rate of the three groups were statistically significant (P< 0.05). The ZNF582 methylation rate in the positive HPV16/18 infection group was 73.7%, while the negative group was 63.9%. Compared with normal tissues, ZNF582 protein was highly expressed in cervical cancer tissues, but mRNA expression was low.
Conclusion: While ZNF582 protein is highly expressed in cervical cancer tissues, it was not sufficient for use as a standard for cervical cancer staging. On the other hand, ZNF582 promoter methylation had high specificity and sensitivity in detecting CINII and highly diseased cervical lesions and could be used as a diagnostic marker for cervical cancer of women.
Keywords: cervical cancer, ZNF582 gene, promoter methylation, HPV16/18, gene expression
Introduction
Cervical cancer is the second most common cancer and also the one of leading causes of cancer death in women in less developed regions.1,2 Xinjiang, especially in the south, has one of the highest incidences of cervical cancer in China. The prevalence and mortality of cervical cancer are both higher in Uygur women than in other ethnic groups in the same region. Surveys suggested that the highest rate of infection in Xinjiang Uygur women is HPV16/18, followed by HPV52, and HPV53.3,4 Although most women are infected with HPV at some point, research has shown that less than 1% of women will eventually progress to cervical cancer.5 Although the diagnostic techniques and treatment strategies for cervical cancer have improved, cervical cancer mortality remains high, and tumor invasion and metastasis remain important obstacles for cervical cancer treatment.6 Therefore, finding an early diagnosis of cervical cancer is crucial.7
Genetic and epigenetic changes in the host and/or viral genome after the continuous infection of the high-risk type of human papillomavirus (hrHPV) are thought to be associated with the development of invasive cervical cancer.8–10 It is known that cervical cancer is caused by the mutation and deletion of genes, but with in-depth exploration of tumors in recent years, there is increasing evidence that hypermethylation and histone modifications of promoter abnormalities are also necessary for cancer tissue metastasis.11 Thus, HPV is sensitive to cervical cancer, but its specificity is low. A large number of false-positive results can lead to unnecessary medical measures and psychological burden.12 Therefore, we need new and better markers with higher specificity and sensitivity for cervical cancer screening.
The progression of HPV-induced precancerous lesions to invasive cancer is due to the accumulation of genetic and epigenetic aberrations affecting host cell genes, leading to activation of oncogenes and/or inactivation of tumor suppressor genes.9 DNA methylation is one of the epigenetic mechanisms that influence gene transcription, chromatin structure, genomic stability, and inactivation of imprinted genes, and X chromosomes.13 When the promoter region of a tumor suppressor gene is highly methylated, this usually results in inactivation of the suppressor gene.14 In some experiments, methylation profiles of human cervical cancer and normal cervix, methylation status in different degrees of cancer tissues and cervical smears, and protein expression were observed, and methylation of the ZNF582 promoter was observed in all cervical cancer samples.15–17 Therefore, we investigated whether gene methylation can be used as a biomarker to examine disease progression and determine whether it can be used as a means of detecting cervical cancer.
Materials and Methods
Patients
We collected 154 cases of fresh cervical tissue specimens from Uygur women obtained by surgical resection or biopsy from May 2010 to May 2018 at the First Affiliated Hospital of Shihezi University, the Third Affiliated Hospital, the Kashgar People’s Hospital and the Xinjiang Autonomous Region Chinese Medicine Hospital (Table S1). The patients had an average age of 47 years (range 23–73), with cervical cancer but had not received chemotherapy or radiotherapy. Lesions included 51 normal cervical tissue or chronic cervicitis cases, 35 cases of CIN (including 10 cases of CINI, 12 cases of CINII, 13 cases of CINIII), and 68 cases of cervical cancer. All samples were confirmed by the hospital pathology department and consent of the patient and her family was obtained. At the same time, we collected the name, age, pathological stage, and other major patient data. The specimen collection was approved by the “Ethics Committee of the First Affiliated Hospital of Shihezi University School of Medicine”, and all patients signed the informed consent.
Methods
Extracting Tissue DNA
Genomic DNA was extracted from 30 mg of tissue with the DNAprep kit. Pure paraffin-embedded tissue DNA is produced in Beijing Tiangen Biotech Co., Ltd. The quality of DNA was assessed by 0.7% agarose gel electrophoresis, and stored DNA samples at −20°C.
Sulfite Modification of DNA
About 1 µg of DNA was subjected to bisulphite treatment using the DNA modification kit (CpGenomeTM, S7820 Kit, Chemicon company). Then, bisulfite-converted DNA was used as a template for detection of ZNF582 gene methylation in all samples using the methylation-specific PCR (MSP) method.
High Risk Human Papillomavirus (HPV16, 18) Infection Detection
Primer design: Optimal primers were designed according to the HPV16 and HPV18 gene sequences provided by the GenBank database on the NCBI website (Table 1). Primers were synthesized by Shanghai Sheng Gong Bioengineering Co., Ltd., and the mass spectrometry results were satisfactory.
 |
Table 1 Primer Information |
PCR detection of HPV16 and HPV18 viral DNA was performed on all samples. The total sample volume of the PCR reaction system is 25 µL, included 12.5 µL PCR mix, 10.5 µL sterile water, primers and DNA. Cycle parameters included: pre-denaturation at 95°C for 10 min, then 35 cycles 95°C 30 s, 57°C 40 s, 72°C 40 s, and a final extension for 5 min at 72°C. The amplified fragment size was 268 bp, the reaction products were run on a 2% agarose gel by electrophoresis, and identified in the gel imaging system (BIO-RAD Gel DocTM XR+ with Image LabTM Software).
Methylation-Specific PCR
Primer design: The ZNF582 gene sequence was obtained from the UCSC website (http://genome.ucsc.edu/), and the methylation-specific PCR primers for ZNF582 were acquired from MethPrimer (http://www.urogene.org/cgi-bin/methprimer/methprimer.cgi) Website Design (Table 1).
RNA Extraction and qPCR
The RNA was extracted from tissue by Trizol. All reagents and solutions used in the procedure were RNase-free. The RNA was further extracted by the RNA extraction kit of RNAprep Pure FFPE Kit (Tiangen Biotech (Beijing) Co., Ltd, DP439). RNA was converted to cDNA by reverse transcriptase. Real-time PCR was performed using the SYBR® Green PCR Kit (Qiagen, Germany) and β-actin as an internal reference gene to detect the expression of the ZNF582 gene, and each sample was tested in three replicates.
Immunohistochemistry
Immunohistochemistry (IHC) was carried out in strict accordance with the second antibody kit instructions. The primary antibody was replaced with phosphate-buffered saline (PBS) as a negative control. The primary antibody (ZNF582 antibody, purchased from Sigma-Aldrich (Shanghai) Trading Co., Ltd.) was diluted to 1:200. The second antibody is a rabbit and mouse universal antibody labeled with horseradish peroxidase (Universal kit (mouse/rabbit polymer method detection system), Beijing Zhongshan Jinqiao Biotechnology Co., Ltd.).
Statistical Analysis
The data were analyzed by the statistical package SPSS 17.0. The χ2- test was used to analyze the qualitative data and correlation. The result data of qPCR detection and measurement data were analyzed by using a two-sample t-test. The P-value <0.05 was considered statistically significant.
Results
Infection of HPV 16 and 18 in Cervical Tissues with Different Degrees of Lesion
Statistical analysis was performed on HPV16 and HPV18 infection in cervical tissues of 154 women. The results showed that in normal and chronic cervicitis, CIN, and cervical cancer tissues, the HPV16 infection rates were 19.6%, 34.3%, and 64.7%, respectively, and demonstrated an increasing trend with severity. The infection rates of HPV18 were 2%, 8.6%, and 13.2%, respectively. The multiple comparison of infection rates of HPV16 in the above groups, and the difference were statistically significant (P<0.05), while that of HPV18 was not. HPV16 and HPV18 infection rates were statistically significant in the CIN and cervical cancer groups, and the difference was also significant in the normal and cervical cancer groups (Figure S1, Table 2).
 |
Table 2 The Infection Status of HPV16/18 |
The Part of Promoter of ZNF582 Gene is Highly Methylated in Cervical Cancer Tissues
We tested the methylation level of the ZNF582 gene promoter in the collected samples. ZNF582 methylation occurred in 9 of 51 normal cervical tissues (17.6%), 16 of 35 CIN tissues (45.7%), and 50 out of 68 cervical cancer cases (73.5%). The difference in methylation rates of the ZNF582 gene in the three groups was statistically significant (P<0.05). With the development of the disease, the probability of methylation showed an increasing trend (Table 3).
 |
Table 3 The Frequency of the ZNF582 Promoter Methylation |
DNA methylation was modified in the ZNF582 genomic promoter region and we designed methylation and non-methylation specific primers. The non-methylated PCR product was 142 bp. A band corresponding to the methylated promoter was present in the sample with methylation, and absent in the unmethylated sample (Figure 1).
Relationship Between Methylation of ZNF582 Gene and HPV Infection in Cervical Cancer Tissues
Among 93 high-grade cervical lesions and cervical cancer tissue samples, 57 HPV16/18 positive and 36 negative samples were included. In the positive group, 42 cases (73.7%) were methylated at the ZNF582 promoter. The negative group had 23 cases (63.9%) of methylation. There was no statistically significant difference between the two groups (χ2 =1.006, P>0.05) (Table 4).
 |
Table 4 The Distribution of ZNF582 Promoter Methylation and HPV16/18 Infection of in CINII, III and Cervical Cancer Tissues |
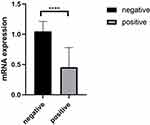
The mRNA Expression Level of ZNF582 Promoter Methylation was Significantly Reduced
The mRNA expression in 10 cases of cervical cancer with methylation was 0.46 ± 0.33, while in 10 cases of normal cervical tissue with no methylation the RNA expression was 1.05 ± 0.16. The expression level of mRNA, in the methylation-positive group, was significantly lower than the negative group (P<0.01), the differences were statistically significant (Table 5, Figure 2).
 |
Table 5 ZNF582 Gene Expression Analysis |
Correlation Between Promoter Methylation of ZNF582 Gene and Clinical Pathological Factors
The results showed that there was no correlation between age, FIGO stage or ZNF582 gene promoter methylation (P>0.05) in 68 cases of cervical cancer (Table 6).
 |
Table 6 ZNF582 Promoter Methylation with Clinical Information of Cervical Cancer Patients |
Diagnostic Efficacy of ZNF582 Promoter Methylation and HPV16/18 Infection
We compared the sensitivity, specificity, and positive and negative predictive value of ZNF582 promoter methylation and HPV16/18 infection detection in normal cervical tissue, low-grade cervical intraepithelial neoplasia (CINI), high-grade cervical intraepithelial neoplasia (CINII and CINIII) and cervical cancer tissue. The results showed that with the sensitivity and specificity of cervical cancer diagnosis, the ZNF582 promoter methylation was 69.9% and 83.6%, respectively. The sensitivity and specificity of detecting the methylation of ZNF582 gene promoter are higher than that of HPV16/18 infection, and the sensitivity and specificity of HPV16/18 detection are 61.3% and 82.0%, respectively (Table 7).
 |
Table 7 Sensitivity and Positive Predictive Value Detection HSIL and Cervical Cancer Tissues, Negative Predictive Value and Specific Analysis of the Normal Tissues and LSIL |
ZNF582 Protein Expression is Increased Expression in Cervical Cancer Tissues
We analyzed ZNF582 protein levels in 87 tissues of cervical cancer tissue and normal cervical tissue by immunohistochemistry. Compared with normal tissues, ZNF582 expression was abnormally upregulated; 86 of 87 cancer patient samples were positive, suggesting ZNF582 protein expression levels can be used to diagnose cervical cancer (Table 8, Figure 3).
 |
Table 8 Statistical Analysis of ZNF582 Expression in Cervical Cancer Tissues and Cervicitis Tissues |
Discussion
Cervical cancer is one of the leading causes of cancer death in women, and the 5-year survival rate of these patients is low.18 Although research in cervical cancer has made great strides, it is still a serious burden, especially in China.19 The overall survival rate of a patient with an early diagnosis of cervical cancer has increased in recent years; however, the prognosis of metastatic cervical cancer remains poor.20 For individualized care and management of patients with cervical cancer, it is important to identify diagnostic and prognostic biomarkers that can be effectively used to guide treatment decisions.7,21,22 Epidemiological and molecular studies support the conclusion that persistent infections of high risk types of human papillomavirus (HPV) are essential but not exclusive prerequisites for cervical carcinogenesis, and has led to the development of HPV DNA testing to improve cervical cancer screening strategies.23,24 Additional genetic and epigenetic alterations are required for progression from precancerous disease to invasive cancer.25,26 Specific epigenetic processes include DNA methylation, chromatin remodeling, histone modification, and microRNA regulations.27 CpG methylation of promoters, an important epigenetic mechanism for gene silencing, is an early and frequently identified alteration in carcinogenesis.28
Immunohistochemical staining showed that ZNF582 was weakly expressed in normal cervical tissues but highly expressed in cervical cancer. However, experimental studies have shown that ZNF582 mRNA has low expression in cervical tissues.
The specific mechanism of the inconsistent expression of ZNF582 at the mRNA and protein levels is unclear, and further experimental verification is required.29–31 It is possible to measure ZNF582 protein expression by immunohistochemistry for cervical cancer diagnosis.
Methylation results showed that in normal tissues, CIN, and cervical cancer tissues, the methylation rates of ZNF582 gene were 17.6%, 45.7%, and 73.5%, respectively. With the severity of disease, the methylation rate of the ZNF582 promoter increased gradually.
At the same time, our study found that there was no significant correlation between age or FIGO stage and the methylation rate of ZNF582 in 68 cases of cervical cancer. We used real-time quantitative PCR to detect ZNF582 gene expression in methylation positive and negative cervical tissues. The results showed that the expression level of the methylation-positive group was significantly lower than the negative group (P<0.01). In this study, we analyzed 93 cases of high-grade cervical injury (CINII and above) and cervical cancer samples, and a total of 57 samples of HPV16/18 infection, 36 cases without infection. The ratio of methylated ZNF582 in the HPV16/18 infection positive group was 73.7%, while the negative was 63.9%. The difference was not statistically significant. This result suggested that HPV16/18 infection may have no relation to the methylation of the ZNF582 gene promoter.
Our results indicated that ZNF582 mRNA was weakly expressed, possibly caused by excessive methylation of the ZNF582 gene, but its protein was highly expressed in cervical cancer tissues, due to other unclear mechanisms in vivo. In the diagnosis of cervical cancer, we compared the sensitivity and specificity of ZNF582 gene methylation and HPV16/18 infection detection. The sensitivity and specificity of detecting the methylation of ZNF582 gene promoter are higher than that of HPV16/18 infection. The methylation rate of the ZNF582 gene promoter was not affected by cervical lesion grade.
Conclusion
In order to reduce the large number of false-positive results of HPV16/18 testing, which may cause unnecessary medical measures and psychological burden in women, the degree of methylation of the ZNF582 gene promoter can be measured. Because of its high specificity and sensitivity to CINII and above, it can be used as a diagnostic indicator of cervical cancer.
Abbreviations
CIN, Cervical intraepithelial neoplasia; HPV, Human papillomavirus; hrHPV, High-risk type of human papillomavirus; MSP, Methylation-specific PCR; ZNF582, Zinc finger protein 582.
Ethical Approval and Consent to Participate
This study was conducted in accordance with the Declaration of Helsinki.
Acknowledgments
We thank International Science Editing for editing this manuscript.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Funding
We acknowledge funding support from the International Science and Technology Collaboration Projector of Xinjiang Production and Construction Corps (grant numbers 2019BC007), the National Natural Science Foundation of China (grant numbers: U1503125, 81660462), the Youth Project Development and Regeneration Key Laboratory of Sichuan Province, the General Project Scientific Research Fund of Chengdu Medical College (grant number: SYS18-07), and the Xinjiang Production and Construction Corps Key Areas Innovation Team Project (grant number: 2018CB002).
Disclosure
The authors declare that they have no competing interests.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Kent A. HPV vaccination and testing. Rev Obstet Gynecol. 2010;3(1):33–34.
3. Wu D, Zhang J, Fan P, et al. Methylation in the promoter regions of WT1, NKX6-1 and DBC1 genes in cervical cancer tissues of Uygur women in Xinjiang. Genet Mol Biol. 2018;41(1):9–17. doi:10.1590/1678-4685-gmb-2016-0146
4. Pan Z, Song Y, Zhe X, et al.Screening for HPV infection in exfoliated cervical cells of women from different ethnic groups in Yili, Xinjiang, China. Sci Rep. 2019;9(1):3468. doi:10.1038/s41598-019-39790-2
5. Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7(1):11–22. doi:10.1038/nrc2050
6. Du Toit GC, Kidd M. Prospective quality of life study of South African women undergoing treatment for advanced-stage cervical cancer. Clin Ther. 2015;37(10):2324–2331. doi:10.1016/j.clinthera.2015.08.018
7. Zhang L, Liu SK, Song L, Yao HR. SP1-induced up-regulation of lncRNA LUCAT1 promotes proliferation, migration and invasion of cervical cancer by sponging miR-181a. Artif Cells Nanomed Biotechnol. 2019;47(1):556–564.
8. Snijders PJ, Steenbergen RD, Heideman DA, Meijer CJ. HPV-mediated cervical carcinogenesis: concepts and clinical implications. J Pathol. 2006;208(2):152–164. doi:10.1002/path.1866
9. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14(6):395–405. doi:10.1038/nrc3728
10. De Strooper LM, Meijer CJ, Berkhof J, et al. Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev Res. 2014;7(12).
11. Fang J, Zhang H, Jin S. Epigenetics and cervical cancer: from pathogenesis to therapy. Tumor Biol. 2014;35(6):5083–5093.
12. Lynge E, Rebolj M. Primary HPV screening for cervical cancer prevention: results from European trials. Nat Rev Clin Oncol. 2009;6(12):699–706. doi:10.1038/nrclinonc.2009.167
13. Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi:10.1101/gad.947102
14. van Splunter AP, Meijer CJLM, Duin S, et al. Host cell deoxyribonucleic acid methylation markers for the detection of high-grade anal intraepithelial neoplasia and anal cancer. Clin Infect Dis. 2018;68(7):1110–1117.
15. Huang R-L, Chang -C-C, Su P-H, et al. Methylomic analysis identifies frequent DNA methylation of zinc finger protein 582 (ZNF582) in cervical neoplasms. PLoS One. 2012;7(7):e41060. doi:10.1371/journal.pone.0041060
16. Li N, He Y, Mi P, Hu Y. ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse: a meta-analysis of related studies in Chinese population. Medicine (Baltimore). 2019;98(6):e14297. doi:10.1097/MD.0000000000014297
17. Liou YL, Zhang TL, Yan T, et al. Combined clinical and genetic testing algorithm for cervical cancer diagnosis. Clin Epigenetics. 2016;8:66. doi:10.1186/s13148-016-0232-3
18. Zhang Y, Li T, Zhang L, et al. Targeting the functional interplay between endoplasmic reticulum oxidoreductin-1alpha and protein disulfide isomerase suppresses the progression of cervical cancer. EBioMedicine. 2019.
19. Torbati MB, Ebrahimian M, Yousefi M, Shaabanzadeh M. GO-PEG as a drug nanocarrier and its antiproliferative effect on human cervical cancer cell line. Artif Cells Nanomed Biotechnol. 2017;45(3):568–573. doi:10.3109/21691401.2016.1161641
20. Dasari S, Wudayagiri R, Valluru L. Cervical cancer: biomarkers for diagnosis and treatment. Clin Chim Acta. 2015;445:7–11. doi:10.1016/j.cca.2015.03.005
21. Somashekhar SP, Ashwin KR. Management of early stage cervical cancer. Rev Recent Clin Trials. 2015;10(4):302–308. doi:10.2174/1574887110666150923113629
22. Cong J, Liu R, Wang X, Jiang H, Zhang Y. MiR-634 decreases cell proliferation and induces apoptosis by targeting mTOR signaling pathway in cervical cancer cells. Artif Cells Nanomed Biotechnol. 2016;44(7):1694–1701. doi:10.3109/21691401.2015.1080171
23. Suba EJ, Michelow PM, Raab SS. Re: human papillomavirus testing in the prevention of cervical cancer. J Natl Cancer Inst. 2011;103(19):
24. Zhu Y, Ren C, Yang L, Zhang X, Liu L, Wang Z. Performance of p16/Ki67 immunostaining, HPV E6/E7 mRNA testing, and HPV DNA assay to detect high-grade cervical dysplasia in women with ASCUS. BMC Cancer. 2019;19(1):271. doi:10.1186/s12885-019-5492-9
25. Soto D, Song C, McLaughlin-Drubin ME. Epigenetic alterations in human papillomavirus-associated cancers. Viruses. 2017;9(9):248. doi:10.3390/v9090248
26. Lattanzio L, Nigro CL. Epigenetics and DNA methylation in cancer. World J Transl Med. 2015;4(01):11–24. doi:10.5528/wjtm.v4.i1.11
27. Saavedra KP, Brebi PM, Roa JC. Epigenetic alterations in preneoplastic and neoplastic lesions of the cervix. Clin Epigenetics. 2012;4(1):13.
28. Yang HJ. Aberrant DNA methylation in cervical carcinogenesis. Chin J Cancer. 2013;32(1):42–48. doi:10.5732/cjc.012.10033
29. Klaes R, Friedrich T, Spitkovsky D, et al. Overexpression of p16(ink4a) as a specific marker for dysplastic and neoplastic epithelial cells of the cervix uteri. Int J Cancer. 2001;92(2):276–284. doi:10.1002/ijc.1174
30. Sano T, Oyama T, Kashiwabara K, Fukuda T, Nakajima T. Expression status of p16 protein is associated with human papillomavirus oncogenic potential in cervical and genital lesions. Am J Pathol. 1998;153(6):1741–1748. doi:10.1016/S0002-9440(10)65689-1
31. Li Y, Nichols MA, Shay JW, Xiong Y. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product pRb. Cancer Res. 1994;54(23):6078–6082.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.