Back to Journals » Drug Design, Development and Therapy » Volume 13
The underlying mechanisms of Jie-Du-Hua-Yu granule for protecting rat liver failure
Authors Qiu H, Mao D, Tang N, Long F, Zhang R, Wang M , Shi Q, Li J , Jiang Q, Chen Y, Wang X
Received 20 July 2018
Accepted for publication 6 December 2018
Published 11 February 2019 Volume 2019:13 Pages 589—600
DOI https://doi.org/10.2147/DDDT.S180969
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sukesh Voruganti
Hua Qiu,1 Dewen Mao,1 Nong Tang,2 Fuli Long,1 Rongzhen Zhang,1 Minggang Wang,1 Qinglan Shi,1 Jiahuan Li,1 Qin Jiang,1 Yueqiao Chen,1 Xiufeng Wang1
1Department of Liver Disease, The First Affiliated Hospital of Guangxi University of Traditional Chinese Medicine, Nanning, Guangxi 530023, China; 2Graduate School, Guangxi University of Chinese Medicine, Nanning, Guangxi 530200, China
Objectives: Jie-Du-Hua-Yu (JDHY) granule is a combination of six traditional Chinese medicines with known therapeutic effect in treating acute liver failure (ALF). The aim of this study was to investigate the amelioration efficacy of JDHY in lipopolysaccharide/D-galactosamine (LPS/D-GalN)-induced ALF in rat and explore the possible molecular mechanism underlying the therapeutic efficacy.
Materials and methods: The efficacy of JDHY was determined by assessing hepatic pathology and function in LPS and D-GalN challenged Wistar rat. We also evaluated the effect of JDHY on LPS-induced Kupffer cells by measuring inflammatory cytokines and determining the phenotypic function. By means of bioinformatics analysis of liver tissue and validation in Kupffer cells, we identified possible pathways involved in the pharmacologic action of mechanism of JDHY.
Results: JDHY could attenuate LPS-induced liver injury in rat by inhibiting apoptosis and increasing hepatic activity. In vitro study showed that JDHY could decrease the production of proinflammatory cytokines (tumor necrosis factor-α, IL6, and interferon-γ), increase anti-inflammatory cytokines (IL10, IL13), and promote cell survival and proliferation, possibly due to inhibition of IκB/nuclear factor-κB (NF-κB) signaling pathway and expression of CD14 and CXCL2, which was consistent with the findings from bioinformatics analysis.
Conclusion: Our results revealed that JDHY protected against LPS-induced liver damage both in vitro and in vivo, by inhibiting the NF-κB-mediated inflammatory pathway, indicating its potential function to treat liver diseases.
Keywords: JDHY, liver failure, NF-κB
Introduction
Acute liver failure (ALF), a rare but life-threatening syndrome, presents a rapid deterioration of liver function and potential multiorgan failure.1,2 With systemic inflammation as the key feature, its poor outcome is strongly correlated with exacerbated systemic inflammatory responses known as cytokine storm. The role of these inflammatory mediators was recently validated in the pathogenesis of ALF by a comprehensive study analyzing 29 different cytokines and chemokines from plasma samples of 522 decompensated cirrhosis patients (with and without ALF) and 40 validated healthy donors.3 Increased levels of proinflammatory cytokines were found in patients with ALF; meanwhile, patients with ALF also present an exacerbated production of anti-inflammatory cytokines such as IL-10.
The mechanisms underlying this intense systemic inflammatory response in ALF are not precisely understood, but recent studies demonstrated that it could be induced by the presence of damage-associated molecular patterns, which could be released by cell damage, and pathogen-associated molecular patterns (PAMPs), such as virus in the systemic circulation.4 In this study, we used lipopolysaccharide (LPS) that is a well-known PAMP. When bound to lipopolysaccharide-binding protein with high affinity, it activates target cells, such as Kupffer, and triggers multiple downstream signaling pathways including promoting nuclear factor-κB (NF-κB) translocation into nucleus and transcription of a battery of proinflammatory cytokines, which in turn further exacerbated liver injury.5,6
Jie-Du-Hua-Yu (JDHY) granule is a traditional Chinese herbal compound, which has been used for over two decades in the treatment of liver diseases. This formula consists of six herbs: Radix paeoniae rubrathe, Artemisia capillaris, Rheum officinale, Radix curcumae, Oldenlandia diffusa, and Acorus gramineus.7,8 Our previous clinical studies suggested that JDHY exhibited protective effect in hepatitis B-related ALF patients and improved liver function.9 In addition, animal experiments demonstrated that JDHY could decrease the expression of caspase-3 mRNA in an ALF model.10 However, the underlying mechanism was poorly understood. Based on previous reports on the beneficial activities of JDHY,7 we hypothesized that JDHY may attenuate LPS-induced liver injury in animal studies. To clarify its protective mechanism, we analyzed the effects of JDHY on LPS-induced damage both in vitro and in vivo. Subsequently, the effort to explore the protective mechanism of JDHY was made by bioinformatical analysis. The gene expression microarray datasets of ALF with and without JDHY from Gene Expression Omnibus (GEO) were generated, and we identified differential expression of genes between groups and performed the functional enrichment analysis. Finally, we confirmed the finding that JDHY could inhibit NF-κB pathway in Kupffer cells, which might play a protective role against inflammatory responses and liver injury.
Materials and methods
Animal studies
Guidelines for the care and use of laboratory animals were followed from IACUC protocol. Male Wistar rats (weight 200±50 g) were purchased from Charles River, Beijing, China. The animals were kept in a room at 24°±2°, with a 12-hour/12-hour light–dark cycle. Before inducing the acute liver injury, rats were fast without diet for 12 hours with no restriction of water. Then rats were randomized into three groups (n=10 per group): control group, ALF group, and ALF group treated with JDHY. In groups with acute hepatic failure, rats were administered with D-galactosamine (D-GalN; 600 mg/kg body weight) and LPS (10 μg/kg body weight) intraperitoneally. In JDHY treatment groups, rats were treated with JDHY by oral gavage at a dose of 4.4 g/kg/d for 2 days before D-gal/LPS challenge as previously described.11 The control rats received saline. After 12 hours postinjection, rats were anesthetized with vaporized isoflurane with airflow of oxygen at the rate of 100–200 mL/min for around 3–5 minutes. Then, blood was collected from orbital venous and separated plasma, serum for alanine aminotransferase (ALT), aspartate aminotransferase (AST), and prothrombin time (PT) biochemistry measurement. Ethics approval for the study was given by The First Affiliated Hospital of Guangxi University of Traditional Chinese Medicine animal experimental ethics committee. National Institutes of Health guide for the care and use of laboratory animals was strictly followed by the authors.
Reagents
JDHY granule was manufactured by Jiangyin Tian Jiang Pharmaceutical Co., Ltd. (Jiangsu, China) and purchased from The First Affiliated Hospital of Guangxi University of Traditional Chinese Medicine. D-GalN (A2795) and LPS (L3023) were purchased from Sigma-Aldrich. The antibody of p53 (Cat#9282), PCNA (Cat#13110), F4/80 (Cat#70076), p-NF-κB (Cat#3033), and p-IκB (Cat#2859) was purchased from CST. pHrodo™ red staining for cell phagocytosis was purchased from Thermo Fisher. Caspase-3 activity kit was purchased from Abcam.
Histological analysis
Liver tissue was obtained immediately after rats were euthanized and then fixed in 4% formalin, followed by embedding in paraffin and were cut into 5 μm serial sections. After deparaffinization and pretreatment with proteinase K, colorimetric TUNEL staining kit (Thermo Fisher Scientific, Waltham, MA, USA) was used to detect apoptotic hepatocytes. Sections were counterstained by hematoxylin and eosin stain.
Gene array analysis
The RNA samples were processed according to manufacturer’s instruction (Agilent Technologies, Santa Clara, CA, USA). In brief, a total of 100 ng RNA was dephosphorylated with calf intestinal alkaline phosphatase and denatured with heat in the presence of dimethyl sulfoxide. T4 RNA ligase was used to ligate cyanine 3-cytidine bisphosphate, a cyanine dye, to dephosphorylated single-stranded RNA (including miRNA). Any unincorporated cyanine dye was removed from samples using MicroBioSpin 6 columns (Bio-Rad Laboratories Inc., Hercules, CA, USA). Purified labeled miRNA probes were then hybridized to 8×15 K mouse miRNA microarrays at 10 rpm for 20 hours at 55°C in a hybridization oven. Next, arrays were washed in Agilent GE Wash Buffer 1 with Triton X-102, and then Agilent GE Wash Buffer 2 with Triton X-102. All slides were scanned at 5 μm resolution using PerkinElmer Scan Array Express array scanner after washing. Images were quantified using Agilent’s Feature Extraction software.
Library preparation and sequencing
Total RNA was extracted using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) from tissue sample following manufacturer’s instructions. RNA samples were incubated with ten units of DNA-free DNase I (Thermo Fisher Scientific) for 30 minutes at 37°C to remove residual genomic DNA. The quality and quantity of purified RNA were measured by the absorbance of 260/280 nm using Nano-drop spectrophotometer (LabTech, Hopkinton, MA, USA). RNA integrity was evaluated by electrophoresis using 1.5% (w/v) agarose gel.
Poly(A) mRNA was isolated with oligo (dT) magnetic beads (Invitrogen) from total RNA samples and fragmented by the RNA fragmentation kit (Ambion). It was used as template for first-strand cDNA synthesis using reverse transcriptase (Invitrogen). The second-strand cDNA was synthesized using RNase H (Invitrogen) and DNA polymerase I (New England Biolabs, Ipswich, MA, USA); 120 bp paired-end cDNA libraries were generated using Illumina Genomic DNA Sample Prep kit (Illumina, USA). The libraries were sequenced on the Illumina HiSeq 2000 of the Rainbow Genomics (Shanghai, China) after loading onto flow cell channels.
Kupffer cell isolation and immortalization
Kupffer cells were isolated from rats (male, Wistar) by collagenase perfusion and followed by differential centrifugation, which was reported before.12 Briefly, livers were perfused with type I buffer (KCl 2.68 mmol/L, NaCl 137 mmol/L, Na2HPO4.12H2O 0.7 mmol/L, glucose 10 mmol/L, HEPES 10 mmol/L, EDTA.Na2 0.5 mmol/L, pH 7.4, with heparin added before use) for 15 minutes and then followed by perfused with type II buffer (KCl 2.68 mmol/L, NaCl 137 mmol/L, Na2HPO4.12H2O 0.7 mmol/L, HEPES 10 mmol/L, glucose 10 mmol/L, CaCl2 5 mmol/L, 0.05% collagenase IV, pH 7.4) for another 15 minutes, and the resulting suspension of liver cells was collected after centrifugation. The pellet was resuspended by 10 mL PBS, adding to a 50 mL tube coated with 50% Percoll and 25% Percoll mixture. After centrifugation at 800 g for 15 minutes at 4°C, there were four bands of components, in which the third band contains most of the Kupffer cells. Collected Kupffer cells, diluting with 15 mL PBS, then resuspended the cell pellet and cultured in incubator.
Primary Kupffer cells were then transfected with SV40 virus expressed from 293 T cells containing PDS152_pLenti-GFP-IRES-SV40-LT and packaging mix plasmids. The immortalized Kupffer cells were validated by different approaches.
Real-time quantitative RT-PCR analysis
miRNA and mRNA expression levels were measured by SYBR-based quantitative PCR (Table S1). The first strand of cDNA PCR template was generated from 50 ng RNA following the manufacturer’s protocols. Approximately 2.5 ng of cDNA was used in the following PCR procedure. Specific RNA level was monitored by 7,900 HT real-time PCR system from Applied Biosystems based on the intensity of SYBR green. Following real-time PCR procedure was used: denaturation (95°C for 10 minutes); amplification and quantification, repeated for 40 cycles (95°C for 15 seconds, 60°C for 30 seconds, and 74°C for 3 seconds with a single fluorescence measurement); and melting curve (from 75°C to 95°C, read every 0.2°C, hold 2 seconds). U6 RNA was used to moralize the miRNA expression, while GAPDH RNA was used to normalize mRNA expression data.
Cell proliferation assay
Colorimetric assay was used to detect cell proliferation using CellTiter 96 AQueous One Solution (MTS) reagent (Promega Corporation, Madison, WI, USA). Cells at a density of 1×104 cells/well were seeded in 96-well plates, followed by culturing at 24-hour intervals for 4 days. After this, a total of 20 μL MTS reagent was added into each well followed by further incubation of 4 hours at 37°C. Absorbance value of each well was measured at 450 nm.
Statistical analysis
All data were demonstrated as mean ± SEM. GraphPad Prism 5 software was used to analyze graphs. Differences among ≥3 groups were compared by one-way ANOVA followed by Tukey post hoc test. Differences between two groups were compared by Student’s t-test. A P-value of <0.05 was defined as statistical significance.
Results
Validation of animal liver failure model
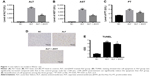
To investigate the function of JDHY for liver injury, we established rat ALF model with D-GalN and LPS in Wistar rats. The levels of ALT, AST, and PT, which are key biomarkers of liver failure, were detected from the blood of rats in each group. Results revealed that rats in ALF group had increased concentration of ALT, AST, and PT compared with the control group. In the group with JDHY pretreatment, the concentration of ALT, AST, and PT was dramatically lower than ALF group (Figure 1A–C). This result indicated that LPS/D-GalN could induce liver injury in Wistar rat, and pretreatment of JDHY could partially protect cell damage induced by LPS/D-GalN. Excessive hepatocyte apoptosis is another key feature of acute liver disease. Thus, therapeutic strategies to inhibit apoptosis in liver injury have the potential to provide a powerful tool for the treatment of liver disease.13 We found that LPS/D-GalN could strongly induce cell apoptosis, which was shown by TUNEL assay. Compared with negative controls, LPS/GaIN led to a 4.5-fold increase in apoptotic cells. However, pretreatment of JDHY only mildly decreased the apoptotic cells (Figure 1D and E).
Microarray and differential genes analysis (GO, KEGG, path-net, gene signal-net)
To interrogate the underlying mechanism of liver failure amelioration by JDHY, mRNA, and miRNA from both ALF and JDHY-pretreated ALF rat liver cells were profiled by microarray analysis using the Affymetrix GeneChip gene Array 1.0 and Affymetrix GeneChip miRNA Array 4.0. A total of 952 mRNAs (fold change >1.5, P<0.05) and 352 miRNAs (fold change >2, P<0.05) with significant difference were identified between the two experimental groups.
First, to identify the biological mechanism correlated with liver failure protection by JDHY, we performed Gene Ontology (GO) analysis using the DAVID program. The top 20 GO categories were shown in Figure 2A, including response to glucocorticoid stimulus, fibrinolysis, regulation of cell shape, and negative regulation of angiogenesis.
Second, to investigate the significant pathways involved in the effect of JDHY, we performed pathway analysis using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database to predict potential functions. The putative genes involved in JDHY function for liver failure relief were enriched in viral carcinogenesis, alcoholism, ether lipid metabolism, and chemokine signaling pathway. The top 20 pathways were listed in Figure 2B.
Third, to interrogate the interactions among significant pathways involving JDHY-induced liver failure amelioration, path-net analysis was performed to demonstrate the pathway interaction. Differential genes were identified as being involved in several key pathways, including cell cycle, p53 signaling pathway, Jak-STAT signaling pathway, and pathways in cancer (Figure 2C).
Fourth, gene signal transduction networks (gene signal-net) were developed to explore gene–gene interaction of differential genes between ALF and ALF-JDHY groups. Results revealed that several genes were identified as core regulators that interact with other molecules in the signaling network, including Cxr2, CD44, Itga6, Ccr2, Ccr5, Adcy10, Cyp3a9, and Pla2g2a (Figure 2D).
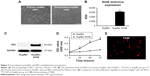
Validation of immortalized rat Kupffer cells (RKCs)
Primary Kupffer cells were isolated from rat and transfected with SV40 virus expressed from 293 T cells containing PDS152_pLenti-GFP-IRES-SV40-LT for immortalization. The RKCs in culture exhibited a typical macrophage-like morphology including membrane ruffling and a heterogeneous content of vacuoles (Figure 3A). The mRNA expression level of SV40 large T antigen in transfected immortalized RKCs was much higher than primary Kupffer cells (Figure 3B). It has been suggested that immortalization by SV40-large T antigen can stabilize cell-cycle regulatory protein p53.14 Our results confirmed this concept and revealed that primary Kupffer cells had undetectable p53 while RKC was detected with high level of p53 protein (Figure 3C). Moreover, RKCs exhibited a stable proliferative capacity, with proliferation of more than two-folds compared with primary Kupffer cells after 72 hours incubation (Figure 3D). The RKCs were strongly positively stained for macrophage cell surface marker F4/80 (Figure 3E). Taken together, these results validated the immortalization of Kupffer cells after SV40 large T antigen transfection in our study.
Effect of JDHY on NF-κB pathway
NF-κB pathway is a well-known inflammation pathway activated by trigger of LPS. From GO analysis, the inflammation response pathway was identified to be important in liver failure ameliorated by JDHY. Activation of NF-κB is followed by expression of a variety of genes, many of which would be expected to participate in inflammatory response.15 Thus, we further interrogated whether NF-κB pathway was involved in the JDHY’s protection from LPS-induced inflammation.
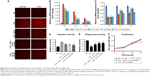
Immunostaining revealed that LPS significantly increased protein level of NF-κB and IκB, which were key elements in NF-κB pathway. JDHY treatment can ameliorate the increase of these two proteins by LPS-induced NF-κB pathway (Figure 4A). LPS was reported to activate Kupffer cells to release Th1 cytokines including tumor necrosis factor-α (TNF-α), IL-6, and interferon-γ (IFN-γ) to cause hepatic injury. Compared with LPS alone, the addition of JDHY in LPS-treated RKCs inhibited the expression of TNF-α, IL6, and IFN-γ, to a similar extent of NF-κB inhibitor. However, combination of JDHY and NF-κB inhibitor did not show a synergistic effect, suggesting that JDHY may function through NF-κB pathway (Figure 4B). Moreover, we observed that JDHY upregulated LPS-induced cytokine stimulation (IL10 and IL13) through non-NF-κB pathway. And IL4 remained unchanged by JDHY in LPS-induced condition (Figure 4C).
Next, we interrogated the involvement of NF-κB pathway in attenuating LPS-induced apoptosis and phagocytosis regulation by JDHY. Caspase 3 activity and phagocytosis results revealed that JDHY can both antagonize LPS-induced apoptosis stimulation and phagocytosis decrease through NF-κB pathway (Figure 4D and E). Meanwhile, results showed that long incubation of LPS led to RKC death, while JDHY can restore the effect of LPS on cell survival by targeting NF-κB pathway (Figure 4F). Taken together, the results indicated that the inhibition of NF-κB pathway played a crucial role in liver failure protection by JDHY.
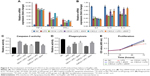
Role of CD14 and CXCL2 in liver failure attenuation by JDHY
Both CD14 and CXCL2 are suggested to be NF-κB pathway responsive genes. Thus, we further studied whether CD14/CXCL2 was involved in JDHY’s protective effect on LPS-induced liver failure. Results showed that the LPS-induced stimulation of TNF-α, IL6, and IFN-γ was inhibited by JDHY, and overexpression of CD14 could partially rescue LPS-induced proinflammation, while addition of JDHY has no add-on effect, suggesting CD14 may be involved. However, overexpression of CXCL2 only has effect in reducing TNF, while it has no effect on IL6 and IFN-γ (Figure 5A). On the contrary, we also profiled some anti-inflammatory factors in this condition. From the results of Figure 5B, we found that IL4 was not effected by JDHY or overexpression of CD14 or CXCL2, while IL10 could be partly restored by JDHY in LPS-induced decrease, which was independent of CD14 or CXCL2 and IL13 may be regulated by CXCL2 but not CD14 (Figure 5B). Moreover, we also observed that JDHY can both antagonize LPS-induced apoptosis stimulation and phagocytosis decrease through CD14 and CXCL2 (Figure 5C and D). The cell survival results revealed that JDHY can protect cells from LPS-induced cell death through CD14 and CXCL2 (Figure 5E).
Discussion
ALF is a highly complicated syndrome featured by strong activation of innate immunity, which leads to sudden shutdown of normal hepatic function and even multiorgan failure. Due to the complexity of the pathogenesis, the clinical management of ALF is very challenging. JDHY granule, which is a combination of six traditional Chinese medicines, is currently being used clinically to treat ALF for many years. Moreover, clinical trials have also shown the protective effects of JDHY granules on ALF.9 In this study, we have provided experimental evidence of the protective role of the JDHY in a rat model of LPS/GaIN-induced liver injury. Combination of LPS and D-GalN strongly induced liver injury, which led to increased serum level of ALT and AST. Short-term treatment of JDHY could partially protect the hepatocyte from injury.
Bioinformatics analysis provided more information to deeply understand the regulation system of JDHY effects in attenuating ALF. NF-κB pathway is a well-known inflammation pathway activated by trigger of LPS. Meanwhile, many studies and clinical therapeutic approaches are also focusing on this pathway to regulate inflammation on ALF. In this study, to validate the findings from bioinformatics study, we used Kupffer cells as our cellular model to validate the findings from bioinformatics. Kupffer cells are a population of tissue macrophages that reside in liver. When activated, cytokines will be released from Kupffer cells and in turn, further activated Kupffer cells forming a positive feedback, which could lead to cytokine storm and systemic complications. Therefore, using Kupffer cell as in vitro model could partly mimic the pathophysiological condition in ALF.
By modulating mRNAs of the different elements on NF-κB pathway or using NF-κB inhibitors, we proved that JDHY has a potential role to decrease the production of proinflammatory cytokines (TNF-α, IL6, and IFN-γ), increase anti-inflammatory cytokines (IL10, IL13), and promote cell survival and proliferation not only in rat model but also on immortalized Kupffer cells. It is noteworthy that Radix curcumae and Rheum officinale, two active gradients of JDHY, were reported to reduce proinflammatory cytokines and reduce inflammation in the serum of ALF mice by blocking NF-κB signal pathways.16,17 Based on our finding and previous reports, we have more confidence that JDHY exerts its clinical effect by targeting NF-κB pathway.
Even the data had showed much evidence to support our hypothesis that JDHY protected against LPS-induced liver damage by inhibiting the NF-κB-mediated inflammatory pathway, indicating its potential to treat liver diseases; there are also many aspects that are uncovered and need further validation. For example, JDHY had very mild efficacy by combining treatment with NF-κB inhibitors. This is possibly because the potency of NF-κB inhibitor is strong enough to inhibit the whole pathway so that there was no space for improvement. Another issue we need to address later is the gap between some mRNA and miRNA modulation; also with the adequate amount of analyzed data, we will further dive in deeper to find potential regulation mechanism in ALF therapeutics. We could also perform the mass spectrometry identification test to narrow down the components in JDHY, which would benefit ALF patients in the future.
Conclusion
JDHY has been identified to be efficacious both in vitro and in vivo. The underlying mechanism of JDHY was probably due to decreasing inflammation biomarkers level, cytokine production secretion levels, cell apoptosis rates, etc, which were mediated by NF-κB pathway. Therefore, JDHY could be used as a potential drug to treat ALF in the future.
Acknowledgments
(1) |
National Natural Science Foundation of China: The mechanism research of the white flower lotus detoxification prescription inhibits hepatitis B virus replication via mir-122 target gene regulatory network (No 81660827). |
(2) |
National Natural Science Foundation of China: The feature analysis of microRNA regulates anti-HBV in hepatocyte endogenous immune signaling network and the intervention mechanism study of white flower lotus detoxification (no 81303066). |
(3) |
National Natural Science Foundation of China: The mechanism research of Jie-Du-Hua-Yu granule regulates NLRP3 inflammatory body activation to antagonism hepatic failure via miRNAs/NF-κB/ROS (No 81774236). |
(4) |
National Natural Science Foundation of China: The disorder characteristic analysis of fulminant hepatic failure pDCs-CTL/CD4+CD25+Treg immune network and the intervention effect of Jie-Du-Hua-Yu granule (No 81460718). |
(5) |
Major projects of Guangxi scientific research and technology development plan: Investigation and analysis of the type of traditional Chinese medicine constitution of patients with chronic hepatitis b in Guangxi and the study on the standardized intervention program of TCM (No 1598011-7). |
Disclosure
The authors report no conflicts of interest in this work.
References
Gustot T, Fernandez J, Garcia E, et al; CANONIC Study Investigators of the EASL-CLIF Consortium. Clinical course of acute-on-chronic liver failure syndrome and effects on prognosis. Hepatology. 2015;62(1):243–252. | ||
Arroyo V, Moreau R, Kamath PS, et al. Acute-on-chronic liver failure in cirrhosis. Nat Rev Dis Primers. 2016;2:16041. | ||
Clària J, Stauber RE, Coenraad MJ, et al; CANONIC Study Investigators of the EASL-CLIF Consortium and the European Foundation for the Study of Chronic Liver Failure (EF-CLIF). Systemic inflammation in decompensated cirrhosis: characterization and role in acute-on-chronic liver failure. Hepatology. 2016;64(4):1249–1264. | ||
Bianchi ME. DAMPs, PAMPs and alarmins: all we need to know about danger. J Leukoc Biol. 2007;81(1):1–5. | ||
Su GL, Klein RD, Aminlari A, et al. Kupffer cell activation by lipopolysaccharide in rats: role for lipopolysaccharide binding protein and Toll-like receptor 4. Hepatology. 2000;31(4):932–936. | ||
Bradham CA, Plümpe J, Manns MP, Brenner DA, Trautwein C. Mechanisms of hepatic toxicity. I. TNF-induced liver injury. Am J Physiol. 1998;275(3 Pt 1):G387–G392. | ||
Wang M, Shi Q, Zhang R, Qiu H, Mao D, Long F. Herbal compound “Jiedu Huayu” reduces liver injury in rats via regulation of IL-2, TLR4, and PCNA expression levels. Evid Based Complement Altern Med. 2017;2017(8):1–9. | ||
Wang M, Qiu H, Zhang R, Long F, Mao D. Subchronic toxicity of herbal compound “Jiedu Huayu” granules in rats. BMC Complement Altern Med. 2017;17(1):450. | ||
Wang N, Wang S, Tang N, Long F, Mao D. [A detoxification and stasis-resolving granules-dominated combination therapy in patients with hepatitis B acute-on-chronic liver failure]. Chin J Integr Tradit West Med Liver Dis. 2014;4:207–209. Chinese. | ||
Mao D, Chen Y, Yu J, Huang G, Wang L, Long F. Effect of Jieduhuayu granule on the expression of Caspase-3mRNA in liver cells of acute liver failure mice. Lishizhen Med Mater Medica Res. 2009;9:2251–2254. | ||
Li G, Qi XP, Wu XY, et al. Verapamil modulates LPS-induced cytokine production via inhibition of NF-kappa B activation in the liver. Inflamm Res. 2006;55(3):108–113. | ||
Kishore R, Hill JR, Mcmullen MR, Frenkel J, Nagy LE. ERK1/2 and Egr-1 contribute to increased TNF-alpha production in rat Kupffer cells after chronic ethanol feeding. Am J Physiol Gastrointest Liver Physiol. 2002;282(1):G6–G15. | ||
Guicciardi ME, Gores GJ. Apoptosis: a mechanism of acute and chronic liver injury. Gut. 2005;54(7):1024–1033. | ||
Peng Y, Murr MM. Establishment of immortalized rat Kupffer cell lines. Cytokine. 2007;37(3):185–191. | ||
O’Neill LA, Kaltschmidt C. NF-kappa B: a crucial transcription factor for glial and neuronal cell function. Trends Neurosci. 1997;20(6):252–258. | ||
Zhang RZ, Qiu H, Wang N, Long FL, Mao DW. Effect of Rheum palmatum L. on NF-κB signaling pathway of mice with acute liver failure. Asian Pac J Trop Med. 2015;8(10):841–847. | ||
Huang X, Lv B, Zhang S, Dai Q, Chen BB, Meng LN. Effects of radix curcumae-derived diterpenoid C on Helicobacter pylori-induced inflammation and nuclear factor kappa B signal pathways. World J Gastroenterol. 2013;19(31):5085–5093. |
Supplementary material
  | Table S1 The gene primers for qPCR |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.