Back to Journals » Infection and Drug Resistance » Volume 14
The Transcription Factor Rv1453 Regulates the Expression of qor and Confers Resistant to Clofazimine in Mycobacterium tuberculosis
Authors Li Y, Fu L, Zhang W, Chen X, Lu Y
Received 11 June 2021
Accepted for publication 27 August 2021
Published 24 September 2021 Volume 2021:14 Pages 3937—3948
DOI https://doi.org/10.2147/IDR.S324043
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Yuanyuan Li, Lei Fu, Weiyan Zhang, Xi Chen, Yu Lu
Beijing Key Laboratory of Drug Resistance Tuberculosis Research, Department of Pharmacology, Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing Chest Hospital, Capital Medical University, Beijing, 101149, People’s Republic of China
Correspondence: Yu Lu
Beijing Key Laboratory of Drug Resistance Tuberculosis Research, Department of Pharmacology, Beijing Tuberculosis and Thoracic Tumor Research Institute, Beijing Chest Hospital, Capital Medical University, Beiguan Street, Tongzhou District, Beijing, People’s Republic of China
Tel +86 10-8950-9358
Email [email protected]
Objective: Clofazimine plays an important role in the treatment of drug-resistant tuberculosis. However, the mechanism of clofazimine resistance remains unclear. In order to slow down the occurrence of clofazimine resistance, it is necessary to study its resistance mechanism.
Methods: In this study, we constructed Rv1453 knockout, complementary and overexpressed strain. The minimum inhibitory concentration (MIC) of clofazimine against Mycobacterium tuberculosis was detected by microplate alamar blue assay (MABA). The transcription levels of Rv1453 and its adjacent genes were detected by quantitative reverse transcriptase PCR. The purified Rv1453 protein was used for electrophoretic mobility shift assay (EMSA) to identify the binding site of Rv1453 protein.
Results: The minimum inhibitory concentration (MIC) of clofazimine increased about 4-fold for the Rv1453 knockout strain and decreased about 4-fold for the Rv1453 overexpressed strain compared with Mycobacterium tuberculosis H37Rv. Further analysis showed that Rv1453 protein, as a regulatory protein, binds to the RNA polymerase binding site of qor and blocks the transcription process.
Conclusion: This study preliminarily revealed that Rv1453 protein of Mycobacterium tuberculosis affects its susceptibility to clofazimine by regulating the transcription level of qor, which is shedding a new light on the mechanism of clofazimine resistance.
Keywords: resistance mechanisms, transcriptional regulation, electrophoretic mobility shift assay, redox
Introduction
Tuberculosis (TB) is a chronic infectious disease caused by the bacillus Mycobacterium tuberculosis and the leading cause of death from a single infectious disease (ranking above HIV/AIDS). Estimated 10 million people worldwide were infected with TB in 2019, and the number of new infections per year has declined slowly in recent years, but not fast enough to reach the target of a 20% reduction between 2015 and 2020. The global treatment success rate of multidrug resistant (MDR)/rifampicin resistant (RR) tuberculosis is only 57%.1 Due to the long course of treatment and high cost of drug-resistant tuberculosis, it brings a heavy burden to families and society. Clofazimine (CFZ) was initially discovered in the development of new antituberculosis drugs. With further studies, it was found that CFZ can also be used for the treatment of drug-resistant TB.2,3 The World Health Organization designated it as group B drug for the treatment of drug-resistant tuberculosis in 2018.4
According to current researches, the mechanism of CFZ resistance is mainly related to the efflux pump. And the reported genes associated with CFZ resistance were Rv0678, Rv1979c and Rv2535c.5–7 The specific mechanism of CFZ has not been fully elucidated. By using a fraction of the membrane isolated from M. smegmatis, it was confirmed that CFZ appeared to compete with menaquinone for electrons. This is the initiating event in the respiratory chain of M. tuberculosis, through which reduced CFZ undergoes spontaneous oxidation to generate reactive oxygen species, such as superoxide and hydrogen peroxide.8 This hypothesis was supported by another study, which claimed that the addition of high concentrations of menaquinone to the culture medium antagonized the antibacterial activity of CFZ.9 Furthermore, inactivating the cytochrome bd-type quinol oxidase in the mycobacterial respiratory chain increased the susceptibility of M. smegmatis to CFZ, which seems to be consistent with inhibition of bacterial respiration.10 The authors speculated that the protective effect of cytochrome bd is mediated by neutralization or inhibition of reactive oxygen species generated by CFZ.
In our previous study, we detected genes associated with clofazimine resistance by whole-genome sequencing from 18 strains of M. tuberculosis. We found that the mutation frequencies of Rv0678, Rv1979c, Rv2535c were 36.4% (4/11), 18.2% (2/11) and 0% (0/11), respectively. However, the mutation frequency of the Rv1453 intergenic region was 54.5% (6/11). In this study, we constructed Rv1453 knockout strain, Rv1453 complementary strain and Rv1453 overexpressed strain to explore the correlation between Rv1453 and clofazimine resistance. The Rv1453 gene encodes the 46.6 KDa transcriptional regulatory protein. And qor gene encodes quinone oxidoreductase, which is adjacent to Rv1453. We speculated that Rv1453-mediated resistance to clofazimine was related to the reduction of reactive oxygen species. The accumulation of quinones in cells could produce a range of toxic effects. Quinones are highly redox active molecules and can generate reactive oxygen radicals during the spontaneous reduction to semiquinone and hydroquinone. Reactive oxygen species (ROS) can cause oxidative damage to DNA, proteins and lipids, and stimulate oxidative stress.11 Quinone oxidoreductases (QOR) have protective and detoxifying effects on cells by reducing quinones to hydroquinone, which could bind to glucuronic acid or sulfate and be excreted. It has been reported that quinone spontaneously loses one electron and becomes semiquinone, whereas in the presence of QOR, quinone can be reduced to hydroquinone by catalyzing the transfer reaction of two electrons, thus avoiding the generation of free radicals.12–14 In this study, we investigated that Rv1453 transcriptional regulatory protein could change the transcriptional level of qor gene to affect the sensitivity of the strain to clofazimine.
Materials and Methods
Bacteria Strains and Culture Conditions
M. tuberculosis H37Rv (ATCC 27294) and recombinant strains were grown at 37°C in 7H9 liquid medium supplemented with 0.2% glycerin, 0.05% Tween 80, and 10% oleic acid-albumin-dextrose-catalase (OADC) or on 7H10 solid medium with 0.5% glycerin and 10% OADC. E. coli strains were grown at 37°C in liquid LB, liquid 2YT or on solid LB medium.
Construction of Recombinant Strains
The Rv1453 knockout strain was constructed as previously reported.15 The Rv1453 gene of M. tuberculosis H37Rv was replaced with res–sacB-hyg–res gene cassette by allelic exchange method. Briefly, chromosomal sequences flanking Rv1453 were amplified by PCR from the genomic DNA of M. tuberculosis H37Rv. Primers LFP and LRP, which contained the Van91I site, respectively, were used to amplify the left arm of Rv1453. Primers RFP and RRP, which contained the Van91I site, respectively, were used to amplify the right arm of Rv1453. These fragments were cloned into the vector p0004s. The resultant plasmid was ligated with the plasmid phAE159 by the PacI site. The phasmid was electroporated into M. smegmatis mc2155 to generate the recombinant phages. M. tuberculosis H37Rv cells were infected with the recombinant phages at 37°C and hygromycin-resistant (HygR) colonies were screened. The Rv1453 overexpressed strain was constructed by electroporation with pMV361 carrying a 1266bp segment of the Rv1453 open reading frame inserted between HindIII and ClaI restriction sites. The Rv1453 complementary strain was constructed by transforming the recombinant pMV361 to the Rv1453 knockout strain. The recombinant strains were verified by RT-PCR. The primers used in this study are listed in Tables S1 and S2.
MIC Determination
The MIC of clofazimine against M. tuberculosis was determined by MABA.16 The concentration of clofazimine ranges from 10 to 0.005 mg/L by using two-fold dilutions method. M. tuberculosis H37Rv cultured to an exponential phase was added to the 96-well plates containing different concentrations of clofazimine. The color of each well was recorded the following day after the addition of Alamar blue and Tween 80. The MIC was defined as the lowest drug concentration that prevented the color change (from blue to pink).
Total RNA Extraction
Total RNA of M. tuberculosis H37Rv and recombinant strains were extracted from freshly cultured bacteria by using the QIAgen RNA mini kit (QIAgen, Germany) according to the manufacturer’s protocol. We used the kit in combination with RNAprotect Bacteria Reagent to ensure reliable gene expression analysis and Lysing Matrix B to achieve complete disruption and homogenization.
Quantitative Reverse Transcriptase PCR
The RNA was reverse transcribed to cDNA using the PrimeScript RT reagent kit with gDNA Eraser (TaKaRa, Japan). The primers listed in Table S1 were used, and the RT-PCR was performed in a 20 μL PCR solution from the TB Green Premix Ex Taq II (TaKaRa, Japan) according to the manufacturer’s protocol. StepOnePlus real-time PCR system (Applied Biosystems, America) was used to perform the RT-PCR with PCR conditions of 95°C for 1 minute, then 40 cycles at 95°C for 5 seconds, 63.4°C for 30 seconds, and 72°C for 30 seconds. The RNA polymerase sigma factor sigA was used as an internal control to normalize the level of target genes for each individual sample. The relative gene expression was calculated using the 2− ∆ ∆Ct method and we used the logarithm of the relative gene expression for the algorithm analysis. The clofazimine drug concentration we used in this experiment was 0.06 μg/mL, and the time points were set as 6 h, 16 h, and 52 h for the dosed group and the blank control group, respectively.
Rv1453 Protein Expression and Purification in E. coli BL21 (DE3)
The Rv1453 gene was amplified by PCR from M. tuberculosis H37Rv genome using the forward and reverse primers, which contained the NdeI and HindIII restriction sites, respectively. We sequenced the DNA fragment cloned into the T vector. We then cloned it to the expression vector pET30a to generate the His tag fusion protein. The recombinant plasmid (pET30a+Rv1453) was transformed into E. coli BL21 (DE3) and grown in 2YT liquid medium at 37°C in an orbital shaker at 200 rpm to an optical density at 600 nm (OD600) of 0.6–0.8. The expression of Rv1453 was induced by the addition of isopropyl–D-1-thiogalactopyranoside at a final concentration of 0.5 mM and grown for an additional 4 h. Cells were harvested by centrifugation at 5000 rpm/min for 5 minutes. His-tagged Rv1453 was purified with Capturem™ His-Tagged Purification Kit (TaKaRa, Japan) according to the manufacturer’s protocol. The protein was purified with 8M urea under denaturing conditions. Purity of the His6-Rv1453 protein was identified by Western blot, and protein concentration was determined by using the BCA Protein Assay Kit (Solarbio, China).
Electrophoretic Mobility Shift Assay (EMSA)
The upstream regions (−500) of Rv1455 and qor gene were amplified by PCR using gene-specific primers. The PCR products were purified by using the MiniBEST DNA segment Purification Kit.
The binding reaction mixture (10×) contained 100 mmol/L Tris-HCl, 500 mmol/L KCl, 10mmol/L DTT, 50% glycerol, 10 mmol/L EDTA. The Rv1453 protein and DNA segment were incubated at 37°C for 1.5 hours. Then, the samples were separated by the native PAGE gel in an ice bath containing 1×Tris-borate-EDTA at 110 V for 1 hour. The native PAGE gel was stained with SYBR Green I dye for about 15 minutes and placed in an ultraviolet analyzer to observe the bands.
We also used the 3ʹbiotin-labeled DNA probes to identify the binding sites. Both the forward and reverse chains of the Rv1455 probe were labeled with biotin, but only the forward chain of the qor probes was labeled with biotin. Electrophoretic mobility shift assays (EMSAs) were carried out according to manufacturer's protocol for the LightShift™ Chemiluminescent EMSA Kit (Thermo Fisher, America). The binding reaction mixture contained 10mmol/L Tris-HCl, 50mmol/L KCl, 1mmol/L DTT, 5% glycerol, 1mmol/L EDTA and 50μg/mL poly (dI-dC). For the Rv1455 probe, the binding reactions of single-chain probe to Rv1453 protein and double-chain probe to Rv1453 protein were detected, respectively.
Computational Analysis
The virtual footprint (http://www.prodoric.de/vfp/) and softBerry (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) software were used to identify the Rv1453 protein-binding motif.
Statistical Analysis
The relative expression levels of Rv1453 gene and its adjacent genes in recombinant strains were compared with M. tuberculosis H37Rv and were tested using a single sample t test. Two-way analysis of variance was used to compare the relative expression levels of various genes among different strains under the action of clofazimine, and the M. tuberculosis H37Rv under the action of clofazimine was taken as a reference. Statistical analysis software was GraphPad prism 8.0 and P < 0.05 was considered statistically different.
Results
Construction and Validation of Recombinant Strains
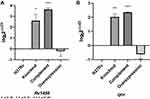
The recombinant plasmid pMV361+Rv1453 and pET30a+Rv1453 has been validated by enzyme digestion and DNA sequencing (Figure S1). Then, we compared the relative expression levels of Rv1453 gene in the Rv1453 knockout strain, Rv1453 complementary strain, Rv1453 overexpressed strain and M. tuberculosis H37Rv to verify the recombinant strains (Figure 1). Compared with the M. tuberculosis H37Rv, the relative expression level of Rv1453 gene in the knockout strain was significantly decreased, while that in the complementary strain and the overexpressed strain were significantly increased. Therefore, it was confirmed that the Rv1453 knockout strain, the Rv1453 complementary strain, and the Rv1453 overexpressed strain were successfully constructed.
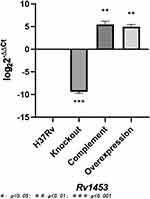 |
Figure 1 The relative expression level of Rv1453 gene in the recombinant strains. |
Susceptibility of Recombinant Strains to Clofazimine
MABA method was used to detect the MIC of isoniazid, rifampicin and clofazimine against M. tuberculosis H37Rv, Rv1453 knockout strain, Rv1453 complementary strain and Rv1453 overexpressed strain (Table 1). We found that, compared with the M. tuberculosis H37Rv, the MIC value of clofazimine to the knockout strain was increased by about 4-fold, while decreased by about 4-fold for the overexpressed strain. As for the complementary strain, it was restored to the same level as that of the M. tuberculosis H37Rv. If hygromycin (75μg/mL) was added to the knockout strain, the MIC of clofazimine against the knockout strain could be restored to the normal level. At the same time, it was found that the MIC of isoniazid and rifampicin could also be increased against the Rv1453 knockout strain, while the MIC of that against complementary strain and overexpressed strain were normal. The MIC of isoniazid and rifampicin against the knockout strain could be reduced to the normal level when added with hygromycin.
 |
Table 1 MIC of Anti-Tuberculosis Drugs Against the Recombinant Strains and H37Rv (μg/ml) |
The Relative Expression Level of Rv1453 Adjacent Genes in the Recombinant Strains
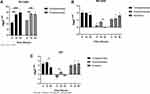
We have found that Rv1453 affects the sensitivity of the strain to clofazimine. Rv1453 encodes the transcriptional regulatory protein, and the relative expression levels of its adjacent genes were detected by RT-PCR to further investigate its transcriptional regulatory function (Figure 2). It was found that the relative expression levels of Rv1455 and qor gene were significantly increased in both the knockout and complementary strains but slightly decreased in the overexpressed strain.
Effects of Clofazimine on the Relative Expression Levels of Rv1453 Gene and Its Adjacent Genes in Recombinant Strains
RT-PCR results showed that the relative expression of Rv1455 gene and qor gene was significantly up-regulated when Rv1453 gene was knockout, while slightly down regulated if Rv1453 gene was overexpressed. In order to further study the effect of clofazimine on the transcription regulation function of Rv1453 protein, the relative expression levels of Rv1453 gene and its adjacent genes in the recombinant strains were detected after clofazimine treatment at 6, 16 and 52 hours, respectively. With the increase in drug treatment time, the relative expression level of Rv1455 gene and qor gene in Rv1453 knockout strain continued to increase slightly, while that decreased in Rv1453 complementary strain and Rv1453 overexpressed strain at 52 h after drug treatment (Figure 3).
Purification and Validation of Rv1453 Protein
In order to further explore how Rv1453 protein plays a negative regulatory role in the transcription of Rv1455 gene and qor gene, we expressed Rv1453 protein in E. coli and purified it, then detected its purity by Western blot (Figure 4). Under denaturation condition, we finally obtained the purified Rv1453 protein and the molecular weight of the protein is approximately 48 kDa.
Transcriptional Regulation Mechanism of Rv1453 Protein on Its Neighboring Genes
According to the predicted sites that may bind to Rv1453 protein, the primers were designed, and the DNA segments of the qor gene promoter region and the Rv1455 gene promoter region were amplified by PCR, respectively. The renatured Rv1453 protein was then used to test whether it could bind to the qor promoter region or the Rv1455 promoter region (Figure 5). It showed that Rv1453 protein could bind to them, and if the amount of protein is too small, the binding reaction is not obvious.
 |
Figure 5 The Rv1453 protein interacts with DNA segments in the qor and Rv1455 promoter regions. |
To explore the DNA binding sites that could bind to Rv1453 protein, two qor double-stranded probes (−325 to −301; −254 to −230) were designed, with biotin-labeled on the forward strand, respectively. We also designed three Rv1455 probes (−176 to −139), one was double-stranded probe with biotin-labeled on both single strands, and the other two were single-stranded biotin-labeled probes. Bands of DNA probe, migration band resulting from the interaction of DNA probe with protein, and decreased migration band resulting from the addition of unlabeled DNA probe were observable in the positive control group, indicating that the experiment was feasible. The migration of bands due to its binding to Rv1453 protein was observed for both binding sites in the promoter region of the qor gene (Figure 6).
 |
Figure 6 Rv1453 protein interacts with DNA segments from the promoter region of the qor gene. |
The interaction between Rv1453 protein and Rv1455 double-stranded probe showed that with the increase in protein concentration, the migration band became more and more obvious, while the single-stranded probe decreased. When the unlabeled Rv1455 double stranded probe was added to the reaction solution, the migration bands decreased and the single stranded probe bands disappeared (Figure 7). Due to the decrease in the single-stranded probe, we further investigated whether Rv1453 protein could bind to single stranded probes. It shows that Rv1455 antisense single-stranded probe can also bind to Rv1453 protein, but that the migration band is weak (Figure 8).
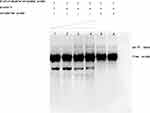 |
Figure 7 Interaction between Rv1453 protein and Rv1455 double-stranded probe. |
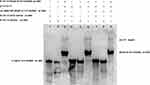 |
Figure 8 Rv1453 protein interacts with single or double strands of Rv1455. |
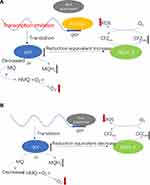
Rv1453 protein has the GGDEF_2 and HTH_30 domain and plays a role in transcriptional regulation. The HTH domain could bind to DNA and be similar to the structure of Bacillus subtilis Pucr transcriptional regulatory protein. The Pucr motif was used to search for binding sites that are located in the promoter region of the Rv1455 gene and qor gene, respectively. We found that the DNA segment that can bind to Rv1453 protein in the promoter region of Rv1455 gene contains the RNA polymerase binding site within qor gene (Figure 9).
Because the qor gene encodes quinone oxidoreductase, which has the function of reducing free radical damage and oxidative stress, we think that Rv1453 protein could change the level of reactive oxygen species by regulating the transcription of qor gene, thus affecting the susceptibility of the M. tuberculosis H37Rv against clofazimine (Figure 10).
Discussion
The purpose of this study was to investigate the relationship between Rv1453 gene and clofazimine resistance in M. tuberculosis. First, the sensitivity of the Rv1453 knockout strain, Rv1453 complementary strain and Rv1453 overexpressed strain to clofazimine were detected by MABA. It was found that the sensitivity of Rv1453 knockout strain to clofazimine was reduced by 4-fold, that of Rv1453 overexpressed strain to clofazimine was increased by 4-fold, and that of Rv1453 complementary strain to clofazimine restored to the same level as M. tuberculosis H37Rv. Therefore, Rv1453 gene is related to the susceptibility of the M. tuberculosis to clofazimine. Besides targeted inhibition, antibiotics can also cause cell death by producing reactive oxygen species.17,18 The phenazine core is the primary structure of CFZ. Since phenazine molecules are auto oxidizable compounds, they can act as electron acceptors, leading to the amount of available ATP decrease.19,20 At the same time, intracellular REDOX enables clofazimine to produce reactive oxygen substances.21 Therefore, the reduced susceptibility of the Rv1453 knockout strain to clofazimine may be due to the decrease in reactive oxygen species. We also found that the MIC of isoniazid and rifampicin could be increased against the Rv1453 knockout strain. Isoniazid and rifampicin can significantly increase the formation of hydroxyl radicals.22,23 In this study, we found that during the culture of Rv1453 knockout strain, the use of hygromycin could restore MIC to normal level. The Rv1453 gene was replaced with a hygromycin resistance gene in the Rv1453 knockout strain, and the hygromycin resistance gene encodes a protein kinase that can phosphorylate hygromycin to make it inactive and thus enable the strain to survive.24 However, ATP was consumed in this process. The increase in reactive oxygen species was related to the increased respiration, higher NADH consumption and the lower ATP/ADP ratio.25 So, we speculate that the drug resistance may be associated with increased ATP and decreased reactive oxygen species.
Compared with M. tuberculosis H37Rv, Rv1453 overexpressed strain was more sensitive to clofazimine, but had no significant change to isoniazid and rifampicin. This is because CFZ can be reduced by NDH-2 and generate reactive oxygen species in its spontaneous oxidation process.8 Reduced quinone reductases lead to increased reduction equivalents, which increase the CFZ’s ability to transfer electrons from NDH-2 to oxygen and lead to increased ROS and oxidative stress.26
Since Rv1453 gene encodes transcriptional regulatory protein, the target genes regulated by Rv1453 gene were further analyzed. It was found that the transcription level of qor and Rv1455 gene in Rv1453 knockout strain and complementary strain were significantly increased, while they were decreased in Rv1453 overexpressed strain. This may be due to the fact that the Rv1453 protein expressed by the plasmid has a weak inhibition on the transcription of Rv1455 and qor genes, while the knockout of Rv1453 had a strong effect on the increase in the transcriptional level of the above two genes. The Rv1455 gene encodes a protein of unknown function and the qor gene encodes quinone reductase. Quinone reductase can directly catalyze the two electrons reduction of quinone to hydroquinone, preventing free radical generation during redox by avoiding the one electron reduction of quinone to the semiquinone.14 And, it has been reported that lpdA gene, which also encodes quinone reductase, can scavenge reactive oxygen species and reduce the oxidative stress in M. tuberculosis.27,28 Inhibition of quinone oxidoreductase activity can increase free radical damage.28 In addition, quinone reductase can transfer reducing equivalents to the electron transport chain, which is important for energy production under anaerobic conditions. The hydroquinone reaches the cell membrane to be oxidized to menaquinone, and along with proton translocation across the membrane, establishes an electrochemical proton gradient or proton motive force that drives ATP synthase to produce ATP, favoring M. tuberculosis survival.29 Since the knockout of Rv1453 gene can increase the transcriptional level of qor, it can reduce the susceptibility of the strain to clofazimine by increasing ATP, reducing ROS and oxidative stress.
Although the transcription level of qor gene in the Rv1453 complementary strain was significantly increased, its sensitivity to clofazimine was consistent with M. tuberculosis H37Rv. This may be related to the action of clofazimine. As shown in Figure 3, with the increase of clofazimine treatment time, the transcription level of Rv1453 in the complementary strain and the overexpressed strain increased significantly. Clofazimine could enhance the transcription level of Rv1453. After the addition of clofazimine, the transcriptional levels of qor gene in the Rv1453 complementary strain decreased significantly at 52H. And, the transcriptional levels of qor gene in the Rv1453 overexpressed strain also decreased, although it was not significant. However, we found no decrease in the Rv1453 knockout strain. It was considered that the Rv1453 knockout strain lacked the transcription regulatory protein encoded by Rv1453 gene, which resulted in the uninhibited transcription of Rv1455 gene and qor gene.
Nucleic acid-binding proteins have traditionally been divided into two categories: DNA-binding proteins and RNA-binding proteins, but recent studies have found a number of proteins that can bind to both DNA and RNA. Proteins that bind nucleic acids in a sequence-specific manner usually contain one or more well-defined structural motifs (zinc finger, leucine zipper, helix-turn-helix, or helix-loop-helix).30 The C-terminal of Rv1453 protein has helix-turn-helix (HTH) domain, which is similar to the structure of Bacillus subtilis Pucr transcriptional regulatory protein, and this domain can bind to target DNA sequence. It has been reported that there is an activation sequence of 5ʹ-WWWCNTTGGTTAA-3ʹ in the upstream of the target gene regulated by PucR, which is called PucR box. So, we predicted the nucleotide sites binding to Rv1453 protein based on the PucR motif and then performed electrophoretic mobility shift assay to study the interaction of Rv1453 protein with the promoter DNA segment of qor gene and Rv1455 gene, respectively. The results showed that Rv1453 protein could bind to the promoter region of Rv1455 gene and qor gene. In order to identify the binding site of Rv1453 protein, biotin labelled probe was used for further study. It is found that Rv1453 protein not only binds to Rv1455 double stranded probe but also binds to Rv1455 single stranded probe (antisense strand). Because Rv1455 gene and qor gene are located on two different strands with adjacent positions and opposite directions, the DNA sequence of Rv1455 single-strand probe (antisense strand) is consistent with a coding sequence of qor gene, and this sequence contains the predicted RNA polymerase binding site. Therefore, Rv1453 protein binding to this site hinders the binding of RNA polymerase, thus affecting the transcription of qor gene. The levels of ROS and ATP in Rv1453 knockout, complementary and overexpressed strains should be detected in further studies to better elucidate the mechanism of Rv1453 mediated clofazimine resistance in M. tuberculosis.
Conclusion
As far as we know, we have confirmed that Rv1453 gene affected the susceptibility of M. tuberculosis to clofazimine for the first time. Further studies showed that the transcriptional regulatory protein encoded by Rv1453 gene could bind to the qor gene sequence that contained the RNA polymerase binding site, thereby inhibiting the transcription process. It was preliminarily revealed that the Rv1453 knockout strain reduced its susceptibility to clofazimine by increasing the transcription level of qor gene, which provided a new idea for further study on the mechanism of clofazimine resistance.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81973367) and Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support (ZYLX202123).
Disclosure
Miss Yuanyuan Li reports grants from National Natural Science Foundation of China, grants from Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support, during the conduct of the study Mr Lei Fu reports grants from National Natural Science Foundation of China, grants from Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support, during the conduct of the study. Mrs Weiyan Zhang reports grants from the National Natural Science Foundation of China, grants from Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support, during the conduct of the study. Mrs Xi Chen reports grants from the National Natural Science Foundation of China, grants from Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support, during the conduct of the study. Professor Yu Lu reports grants from the National Natural Science Foundation of China, grants from Beijing Hospitals Authority Clinical Medicine Development of Special Funding Support, outside the submitted work. The authors report no conflicts of interest in this work.
References
1. World Health Organization. Global tuberculosis report 2020. Geneva: World Health Organization; 2020.
2. Tang S, Yao L, Hao X, et al. Clofazimine for the treatment of multidrug-resistant tuberculosis: prospective, multicenter, randomized controlled study in China. Clin Infect Dis. 2015;60(9):1361–1367.
3. Xu HB, Jiang RH, Xiao HP. Clofazimine in the treatment of multidrug-resistant tuberculosis. Clin Microbiol Infect. 2012;18(11):1104–1110. doi:10.1111/j.1469-0691.2011.03716.x
4. World Health Organization. Global tuberculosis report; 2018.
5. Hartkoorn RC, Uplekar S, Cole ST. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58(5):2979–2981. doi:10.1128/AAC.00037-14
6. Zhang S, Chen J, Cui P, Shi W, Zhang W, Zhang Y. Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother. 2015;70(9):2507–2510. doi:10.1093/jac/dkv150
7. Almeida D, Ioerger T, Tyagi S, et al. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2016;60(8):4590–4599. doi:10.1128/AAC.00753-16
8. Yano T, Kassovska-Bratinova S, Teh JS, et al. Reduction of clofazimine by mycobacterial type 2 NADH: quinone oxidoreductase: a pathway for the generation of bactericidal levels of reactive oxygen species. J Biol Chem. 2011;286(12):10276–10287. doi:10.1074/jbc.M110.200501
9. Lechartier B, Cole ST. Mode of action of clofazimine and combination therapy with benzothiazinones against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2015;59(8):4457–4463. doi:10.1128/AAC.00395-15
10. Lu P, Heineke MH, Koul A, et al. The cytochrome bd-type quinol oxidase is important for survival of Mycobacterium smegmatis under peroxide and antibiotic-induced stress. Sci Rep. 2015;5:10333. doi:10.1038/srep10333
11. Bolton JL, Trush MA, Penning TM, Dryhurst G, Monks TJ. Role of quinones in toxicology. Chem Res Toxicol. 2000;13(3):135–160. doi:10.1021/tx9902082
12. Oppermann U. Carbonyl reductases: the complex relationships of mammalian carbonyl- and quinone-reducing enzymes and their role in physiology. Annu Rev Pharmacol Toxicol. 2007;47:293–322. doi:10.1146/annurev.pharmtox.47.120505.105316
13. Porté S, Valencia E, Yakovtseva EA, et al. Three-dimensional structure and enzymatic function of proapoptotic human p53-inducible quinone oxidoreductase PIG3. J Biol Chem. 2009;284(25):17194–17205. doi:10.1074/jbc.M109.001800
14. Siraki AG, Klotz LO, Kehrer JP. Free radicals and reactive oxygen species. Compr Toxicol. 2018;10:262–294.
15. Bardarov S, Bardarov S
16. Xu J, Wang B, Hu M, et al. Primary clofazimine and bedaquiline resistance among isolates from patients with multidrug-resistant tuberculosis. Antimicrob Agents Chemother. 2017;61(6). doi:10.1128/AAC.00239-17.
17. Belenky P, Ye JD, Porter CB, et al. Bactericidal antibiotics induce toxic metabolic perturbations that lead to cellular damage. Cell Rep. 2015;13(5):968–980. doi:10.1016/j.celrep.2015.09.059
18. Dwyer DJ, Belenky PA, Yang JH, et al. Antibiotics induce redox-related physiological alterations as part of their lethality. Proc Natl Acad Sci U S A. 2014;111(20):E2100–E2109. doi:10.1073/pnas.1401876111
19. Heikal A, Hards K, Cheung C-Y, et al. Activation of type II NADH dehydrogenase by quinolinequinones mediates antitubercular cell death. J Antimicrob Chemother. 2016;71(10):2840–2847. doi:10.1093/jac/dkw244
20. Murugesan D, Ray PC, Bayliss T, et al. 2-Mercapto-Quinazolinones as inhibitors of type II NADH dehydrogenase and Mycobacterium tuberculosis: structure–activity relationships, mechanism of action and absorption, distribution, metabolism, and excretion characterization. ACS Infect Dis. 2018;4(6):954–969. doi:10.1021/acsinfecdis.7b00275
21. Cholo MC, Mothiba MT, Fourie B, Anderson R. Mechanisms of action and therapeutic efficacies of the lipophilic antimycobacterial agents clofazimine and bedaquiline. J Antimicrob Chemother. 2017;72(2):338–353. doi:10.1093/jac/dkw426
22. Zeng S, Soetaert K, Ravon F, et al. Isoniazid bactericidal activity involves electron transport chain perturbation. Antimicrob Agents Chemother. 2019;63(3). doi:10.1128/AAC.01841-18.
23. Piccaro G, Pietraforte D, Giannoni F, Mustazzolu A, Fattorini L. Rifampin induces hydroxyl radical formation in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2014;58(12):7527–7533. doi:10.1128/AAC.03169-14
24. van den Elzen PJ, Townsend J, Lee KY, Bedbrook JR. A chimaeric hygromycin resistance gene as a selectable marker in plant cells. Plant Mol Biol. 1985;5(5):299–302. doi:10.1007/BF00020627
25. Iqbal IK, Bajeli S, Akela AK, Kumar A. Bioenergetics of mycobacterium: an emerging landscape for drug discovery. Pathogens. 2018;7(1):24.
26. Lamprecht DA, Finin PM, Rahman MA, et al. Turning the respiratory flexibility of Mycobacterium tuberculosis against itself. Nat Commun. 2016;7:12393. doi:10.1038/ncomms12393
27. Zheng H, Lu L, Wang B, et al. Genetic basis of virulence attenuation revealed by comparative genomic analysis of Mycobacterium tuberculosis strain H37Ra versus H37Rv. PLoS One. 2008;3(6):e2375. doi:10.1371/journal.pone.0002375
28. Akhtar P, Srivastava S, Srivastava A, Srivastava M, Srivastava BS, Srivastava R. Rv3303c of Mycobacterium tuberculosis protects tubercle bacilli against oxidative stress in vivo and contributes to virulence in mice. Microbes Infect. 2006;8(14–15):2855–2862. doi:10.1016/j.micinf.2006.09.004
29. Sellamuthu S, Singh M, Kumar A, Singh SK. Type-II NADH dehydrogenase (NDH-2): a promising therapeutic target for antitubercular and antibacterial drug discovery. Expert Opin Ther Targets. 2017;21(6):559–570. doi:10.1080/14728222.2017.1327577
30. Bartas M, Červeň J, Guziurová S, Slychko K, Pečinka P. Amino acid composition in various types of nucleic acid-binding proteins. Int J Mol Sci. 2021;22(2):922. doi:10.3390/ijms22020922
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.