Back to Journals » Diabetes, Metabolic Syndrome and Obesity » Volume 15
The Single Nucleotide Polymorphisms (rs1292037 and rs13137) in miR-21 Were Associated with T2DM in a Chinese Population
Authors Li Y , Yang J, Tao W, Yang M, Wang X, Lu T, Li C, Yang Y, Yao Y
Received 22 October 2021
Accepted for publication 24 December 2021
Published 20 January 2022 Volume 2022:15 Pages 189—198
DOI https://doi.org/10.2147/DMSO.S345758
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Juei-Tang Cheng
Yiping Li,1,* Jia Yang,2,* Wenyu Tao,1 Man Yang,1 Xiaoling Wang,1 Tinglian Lu,1,3 Chuanyin Li,2 Ying Yang,1 Yufeng Yao2
1Department of Endocrinology, The Affiliated Hospital of Yunnan University & The Second People’s Hospital of Yunnan Province, Kunming City, Yunnan, People’s Republic of China; 2Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming City, People’s Republic of China; 3School of Clinic Medicine, Dali University, Dali City, Yunnan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ying Yang
Department of Endocrinology, The Affiliated Hospital of Yunnan University & The Second People’s Hospital of Yunnan Province, Kunming, 650021, Yunnan, People’s Republic of China
Email [email protected]
Yufeng Yao
Institute of Medical Biology, Chinese Academy of Medical Sciences & Peking Union Medical College, Kunming, Yunnan, 650118, People’s Republic of China
Email [email protected]; [email protected]
Background: Insulin receptor (INSR), insulin receptor substrate (IRS) and glucose transporter 4 (GLUT4) play important roles in the insulin resistance pathway. The microRNA (miRNA or miR) involved in INSR, IRS or GLUT4 could be associated with the development of type 2 diabetes (T2DM).
Methods: The aim of this study was to investigate the association of T2DM with 12 single nucleotide polymorphisms (SNPs) in 7 miRNAs (miR-195, miR-126, miR-144, miR-155, miR-21, miR-93 and miR-222) involved in the insulin resistance pathway. A total of 1593 subjects with T2DM and 1656 nondiabetic subjects were genotyped. Then, the associations of these SNPs with the development of T2DM and individual metabolic traits were evaluated, such as fasting plasma glucose (FPG) and glycosylated haemoglobin (HbA1C).
Results: Our data showed that the C allele of rs1292037 in miR-21 could increase the risk of developing T2DM (P = 0.002, OR = 1.17; 95% CI: 1.06– 1.29). In addition, the T allele of rs13137 in miR-21 could be a risk factor for T2DM (P = 0.003, OR = 1.16; 95% CI: 1.05– 1.28). According to inheritance mode analysis, compared with the T/T-T/C genotype, the C/C genotype of rs1292037 showed a risk effect in T2DM in the recessive mode (P = 0.001, OR = 1.35; 95% CI: 1.13– 1.63). For rs13137, compared with the A/A-A/T genotype, the T/T genotype also showed a risk effect in T2DM in the recessive mode (P = 0.001, OR = 1.35; 95% CI: 1.13– 1.62). Moreover, in the nondiabetic group, compared with the rs78312845 A/G (FPG = 5.177± 0.488mmol/L; HbA1C = 5.147± 0.293%) and A/A genotypes (FPG = 5.155± 0.486mmol/L; HbA1C = 5.136± 0.299%), the G/G genotype (FPG = 4.887± 0.482mmol/L; HbA1C = 4.960± 0.397%) was associated with lower FPG (P = 0.012 and 0.019) and HbA1C (P = 0.008 and 0.011).
Conclusion: Our results revealed that rs1292037 and rs13137 in miR-21 were associated with T2DM susceptibility in a Han Chinese population. Moreover, the rs78312845 in miR-195 contributed to the level of FPG and HbA1C in nondiabetic group in the Han Chinese population.
Keywords: polymorphisms, T2DM, insulin resistance, Chinese population, microRNA
Introduction
Diabetes mellitus (DM) has received attention due to its high global prevalence and its severe complications, which lead to high mortality and disability. In China, approximately 140.9 million people had diabetes in 2021, which has the largest numbers of adults with diabetes aged 20–79 years.1 Type 2 diabetes (T2DM), which accounts for 90–95% of all diabetes cases, is triggered by insulin resistance in peripheral tissues.2
Adipose tissue and the liver are considered the major insulin-responsive tissues.3 In adipocytes and hepatocytes, insulin receptor (INSR), insulin receptor substrate (IRS) and glucose transporter 4 (GLUT4) are involved in one of the classic insulin resistance signalling pathways.3,4 The binding of insulin to INSR stimulates IRS and triggers protein kinase C (Akt), and the translocation of GLUT4 promotes the uptake of glucose. The impairment of any molecule in insulin signalling could have detrimental effects on insulin action, therefore leading to insulin resistance and T2DM.3,4
MicroRNAs (miRNAs or miRs), which are derived from endogenous hairpin structured transcripts of the genome, contribute to mRNA posttranscriptional regulation.5 The differential expression of miRNAs among diseases provides evidence that miRNAs play an important role in diseases, such as T2DM.6–14 The miR-195 targeted the INSR in hepatocyte and was upregulated in the hyperglycemic rat liver.15,16 The miR-126 and miR-144 targeted the IRS in adipocyte and were regulated in T2DM subjects.6,9,12,17–19 The miR-155 impaired the insulin sensitivity by targeting INSR and GLUT4 in adipocyte, and lower expression in men with obesity.20,21 The miR-21, miR-222 and miR-93 were proved to target the GLUT4 or its translocation in adipocyte.11,22,23 The expression of miR-21 was shown to be decreased in T2DM subjects,13 obese T2DM subjects,14 obese subjects10,14 and metabolic syndrome subjects.7 The miR-222 was upregulated in the hyperglycemic rat adipose tissue and the miR-222 expression was increasing in response to hyperglycemia in 3T3-L1 adipocyte.15 The miR-93 expression was upregulated in polycystic ovary syndrome with intrinsic insulin resistance, and in non-polycystic ovary syndrome women with insulin resistance.11
Moreover, single nucleotide polymorphisms (SNPs) in miRNAs could have an effect on not only the expression of miRNAs but also target gene mRNA degradation, translation and expression,5,24 which could cause diseases, such as T2DM.25 Therefore, SNPs in miRNAs that regulate insulin resistance by targeting INSR, IRS, Akt and GLUT4 not only change the quantity and quality of microRNAs but also influence the binding between microRNAs and target genes, which are involved in insulin resistance in adipocytes and hepatocytes.11,16–23 For example, in 2019, Zhang et al reported that the rs13137T/T genotype had lower miR-21 levels (P=0.027), and such lower miR-21 level was associated with T2DM, obese nondiabetic and metabolic syndrome subjects.7,13,14,26
In 2019, we investigated the twelve SNPs in 9 miRNAs (rs10459194 in miR-135a-2, rs10993081 and rs7045890 in let-7d, rs2296616 in miR-107, rs2402959 and rs6965643 in miR-96, rs24168 in miR-29a, rs3745453 in miR-23a, rs4636297 in miR-126, rs8089787 and rs9948906 in miR-133a-1 and rs999885 in miR-106b) in 784 T2DM subjects and 846 no-diabetic subjects, and our results have identified the associations of the rs3745453 of miR-23a targeting to GLUT4 and the rs2402959 of miR-96 targeting to IRS with T2DM.27 Therefore, in the current study, we investigated the associations of 12 SNPs in 7 miRNAs (rs78312845 in miR-195, rs2297537 and rs2297538 in miR-126, rs28448745 in miR-144, rs1547354 in miR-155, rs1292037 and rs13137 in miR-21, rs1527423 in miR-93 and rs2858061, rs34678647, rs2858060 and rs2745709 in miR-222) with T2DM in a Han Chinese population. Moreover, we evaluated the association of these SNPs with individual metabolic traits in the nondiabetic (NDM) group.
Materials and Methods
Ethics Statement
This study was approved by the Institutional Review Boards of the Affiliated Hospital of Yunnan University and the Ethical protocol approval number was 2016043. The protocol used in this investigation was in accordance with the principles expressed in the 1975 Declaration of Helsinki, which was revised in 2008. All participants provided written informed consent.
Subjects
A total of 1593 subjects (990 males and 603 females) who were diagnosed with T2DM at the affiliated Hospital of Yunnan University from January 2017 to November 2018 were recruited into this study. The diagnosis of T2DM was in accordance with the World Health Organization criteria published in 1999 and American Diabetes Association (ADA) guidelines in 2017.2 In brief, the criteria for inclusion of T2DM group was the T2DM subjects with fasting plasma glucose ≥7.0mmol/L or 2-hours postprandial plasma glucose≥11.1mmol/L or a random plasma glucose≥11.1mmol/L. The criteria for exclusion of T2DM group was: ①The islet beta-cell function was evaluated by measurement of fasting insulin and C peptides level and glucose-load insulin and C peptides level to exclude subjects with type 1 diabetes; ②The islet cell autoantibodies and glutamic acid decarboxylase autoantibodies were detected to exclude subjects with latent autoimmune diabetes in adults;28 ③Gestational women were not included; ④Specific types of diabetes were excluded by history illness, such as glucocorticoid induced diabetes. The NDM group included 1656 subjects (1048 males and 608 females) with no family history of diabetes mellitus who were undergoing routine health check-ups at the affiliated Hospital of Yunnan University. Subjects with diabetes were excluded from the NDM group. In addition, subjects with prediabetes were excluded from the NDM group (subjects with fasting plasma glucose (FPG) greater than 6.1 mmol/L and/or glycosylated haemoglobin (HbA1C) greater than 5.7%). In addition, subjects with hypertension or coronary heart disease were excluded from the NDM group. All participants (T2DM and NDM) self-reported being ethnically Han.
Laboratory Measurements
Venous blood samples were collected in the morning after the subjects had fasted for 12 hours. Fasting plasma glucose (FPG) was assayed using the glucose oxidase method. Total cholesterol (TC), high-density lipoprotein cholesterol (HDL-C), triglycerides (TG) and low-density lipoprotein cholesterol (LDL-C) were determined by enzymatic methods. HbA1C was determined by immunoturbidimetry. All laboratory measurements were performed on a HITACHI 7600–020 Automatic Analyzer.
miRNA Selection and SNP Genotyping
A total of twelve SNPs in the seven miRNAs (miR-195, miR-126, miR-144, miR-155, miR-21, miR-93 and miR-222) were selected for genotyping, as the miRNAs play an important role in the insulin resistance pathway by directly targeting INSR, IRS or GLUT4. Genomic DNA was obtained from EDTA anticoagulated whole blood of the subjects using a QIAamp Blood Mini Kit. The probes and primers used for genotyping were all purchased from ABI (http://www.appliedbiosystems.com). The twelve SNPs were genotyped using the TaqMan fluorescent quantitative PCR method with the QuantStudio™ Real-Time PCR instrument. The total PCR volume was 5 µL, and the reaction conditions were 95 °C predenaturation for 10 minutes, 40 cycles of 95 °C denaturation for 15 seconds, 60 °C annealing for 1 minute, and finally 60 °C extension for 5 minutes. Deionized water was used to replace template DNA as a negative control. The data were analysed by TaqMan Genotyper Software (Version 1.3.1). To identify the accuracy of SNP genotyping using the TaqMan assay, samples with each genotype of the twelve SNPs were sequenced. Then, different genotypes samples were used as positive sample in each TaqMan assay.
Statistical Analysis
The age and glucose and lipid parameters of the subjects enrolled in the present study are expressed as the mean±standard deviation. The analyses of the age, sex and clinical metabolic parameter differences between the T2DM and NDM groups were calculated. The Hardy-Weinberg equilibrium (HWE) of each SNP in both the T2DM and NDM groups, the allele and genotype frequencies of the SNPs, the differences in allele and genotype frequencies between the T2DM and NDM groups and the odds ratios (ORs) with associated 95% confidence intervals (CIs) of allele-specific risks were calculated by SHEsis software.29,30 The association between each SNP and T2DM was analysed for mode of inheritance using SNPStats after adjusting the sex and age.31 Linkage disequilibrium (LD) measures involving these SNPs for all individuals in the current study were calculated using SHEsis.29,30 If there is a strong linkage between SNPs (D’ > 0.7), the frequencies of the haplotypes constructed using these SNPs were calculated, and association of haplotypes with T2DM were evaluated.29,30 The Akaike information criterion (AIC) and Bayes information criterion (BIC) were used to determine the best fit model for each SNP. Bonferroni correction was performed for multiple comparisons, and the significance threshold was set at P<0.004 (0.05/12). Analysis of variance (ANOVA) was used to compare the differences in metabolic traits between the three genotype groups and these 12 SNPs in the NDM group. Bonferroni correction was used to compare the differences in metabolic parameters between three genotypes in the NDM group, and the significance threshold was set at P<0.05. Power-analysis was performed using power and sample size calculations.32 Statistical analyses were performed using SPSS 21 (Chicago, IL) and GraphPad Prism 7.00.
Results
Subject Characteristics
Table 1 lists the clinical characteristics and glucose and lipid metabolic parameters of the enrolled subjects. The age of the T2DM group was 58.014±12.233 years old (male 57.726±12.497 and female 58.488±11.781, respectively) and was 58.192±11.543 years old (male subjects 58.385±11.405 and female 57.860±11.780, respectively) in the NDM group. In T2DM, there was 990 males and 603 females. In the NDM group, there was 1048 males and 608 females. There were no age or sex differences between the T2DM and NDM groups. In NDM group, the TC, HDL-C, TG, LDL-C, FPG, and HbA1C were 4.897±1.020mmol/L, 1.712±1.149mmol/L, 1.339±0.355mmol/L, 3.080±0.890mmol/L, 5.156±0.487mmol/L and 5.135±0.300%. In T2DM group, the TC, HDL-C, TGs, LDL-C, FPG, and HbA1C were 4.817±1.118mmol/L, 2.458±2.085mmol/L, 1.085±0.284mmol/L, 2.808±0.959mmol/L, 8.136±2.624mmol/L and 9.064±2.703%. Significant differences were observed in glucose and lipid metabolic parameters (TC, HDL-C, TGs, LDL-C, FPG, and HbA1C) between the T2DM and NDM groups (Table 1). Specifically, the T2DM group had lower TC and LDL-C than the NDM group, as antihyperlipidemic agents were prescribed at the diagnosis of T2DM.
 |
Table 1 Clinical Characteristics and Glucose and Lipid Metabolic Parameters of the Subjects Enrolled in the Present Study (Data are the Mean ± SD) |
Association of the Twelve SNPs with T2DM
The allele and genotype frequencies for 8 SNPs in miRNAs located in autosomal chromosomes are listed in Table 2. The genotype frequencies for SNPs were in HWE for the T2DM and NDM groups (P>0.05), except for rs1547354 in the T2DM group (P=0.05). As shown in Table 2, rs78312845 in miR-195, rs2297537 and rs2297538 in miR-126, rs28448745 in miR-144, rs1547354 in miR-155 and rs1527423 in miR-93 showed no association with T2DM (P>0.05). However, the allelic and genotypic distribution of rs1292037 and rs13137 in miR-21 showed significant difference between the NDM and T2DM groups (P<0.004). The C allele of rs1292037 in miR-21 could increase the risk of developing T2DM (P=0.002, OR=1.17; 95% CI: 1.06–1.29), and the T allele of rs13137 in miR-21 could also be a risk factor for T2DM (P=0.003, OR=1.16; 95% CI: 1.05–1.28) (Table 2). For miR-222 located on the X chromosome, the allele and genotype frequencies of four SNPs (rs2858061, rs34678647, rs2858060 and rs2745709) in miR-222 in the T2DM female and NDM female groups are listed in Table 3. The four SNPs in miR-222 showed no association with T2DM (P>0.05).
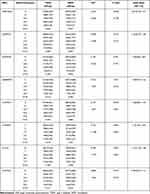 |
Table 2 Comparison of Genotypic and Allelic Distribution of Eight SNPs Among Six microRNAs Between T2DM Group and NDM Group |
 |
Table 3 Comparison of Genotypic and Allelic Distribution of Four SNPs in microRNA-222 Between T2DM Group and NDM Group in Female Subjects |
Mode of Inheritance Analysis
Table 4 and Supplement Table 1 present the results of analyses conducted to determine the mode of inheritance for the twelve SNPs. To compare each inheritance model (codominant, dominant, recessive, overdominant and log-additive) to the most general model, the AIC and BIC were calculated to identify the inheritance model that best fit the data.31 The model with the smallest AIC and BIC values corresponds to the minimal expected entropy.31 The best fit inheritance model with the lowest AIC for rs1292037 and rs13137 in miR-21 was recessive. For rs1292037, the C/C genotype showed a risk effect in T2DM (P=0.001, OR=1.35; 95% CI: 1.13–1.63). For rs13137, the T/T genotype showed a risk effect in T2DM (P=0.001, OR=1.35; 95% CI: 1.13–1.62).
 |
Table 4 Different Inheritance Models of rs1292037 and rs13137 in microRNA-21 Between NDM Group and T2DM Group |
Association of the Haplotypes of the miR-126, miR-21 and miR-222 Gene SNPs with T2DM
According to the LD value (D’≥0.7), we constructed the haplotypes in miR-126, miR-21 and miR-222. For rs2297537 and rs2297538 in miR-126, the LD value was 0.963 for rs2297537-rs2297538. For rs1292037 and rs13137 in miR-21, the LD value was 0.995 for rs1292037-rs13137. For rs34678647, rs2745709 and rs2858060 in the miR-222, the LD value was 0.714 for rs34678647 and rs2858060, 0.889 for rs34678647 and rs2745709, and 0.984 for rs2745709 and rs2858060. Our results showed that only rs1292037-rs13137 haplotype in miR-21 was associated with T2DM. The rs1292037C-rs13137T haplotype was a risk factor for the development of T2DM (P=0.003, OR=1.16; 95% CI:1.05–1.28) and the rs1292037T-rs13137A haplotype was a protective factor of T2DM (P=0.003, OR=0.86; 95% CI:0.78–0.95) (Table 5). The rs2297537-rs2297538 and rs34678647-rs2745709-rs2858060 haplotypes were not associated with the T2DM (Data not shown).
 |
Table 5 Haplotype-Analysis of rs1292037-rs13137 in the microRNA-21 Between T2DM Group and NDM Group |
Association Between Genotype and Metabolic Traits
In the NDM group, compared with the A/G genotype, rs78312845 in the miR-195G/G genotype was associated with lower FPG (P=0.012) and HbA1C (P=0.008) after Bonferroni correction (Figure 1). Similarly, compared with the A/A genotype, rs78312845 in the miR-195 G/G genotype was also associated with lower FPG (P=0.019) and HbA1C (P=0.011) after Bonferroni correction (Figure 1). No significant associations between the other eleven SNP genotypes were observed with metabolic traits in the NDM groups (data not shown).
Discussion
The association of SNPs in miRNAs with diseases has been widely studied in different diseases, such as cancer, metabolic diseases and sepsis. Here, based on the key role of miRNAs targeting INSR/IRS/GLUT4 in hepatocytes and adipocytes, we investigated the associations of 12 SNPs in 7 miRNAs that mediated in the insulin resistance pathway by targeting INSR, IRS or GLUT4 and found that rs1292037 and rs13137 in miR-21 were associated with T2DM susceptibility in a Han Chinese population.
The overexpression of miR-21 has been reported to improve insulin sensitivity by increasing the phosphorylation of Akt in insulin-resistant adipocytes, therefore increasing GLUT4 translocation.22 In addition, miR-21 was aberrantly downregulated or decreased in person with T2DM, obese nondiabetic and metabolic syndrome subjects.7,13,14 These results indicated that lower miR-21 expression was associated with T2DM. In the current study, we found that the T allele and T/T genotype of rs13137 could be risk factors for the development of T2DM. In 2019, Zhang et al reported that individuals with the rs13137T/T genotype in miR-21 had lower miR-21 levels in plasma than individuals with the rs13137A/A genotype.26 Thus, the reason the rs13137 T allele and T/T genotype were associated with T2DM in the current study could be that rs13137 influences the level of miR-21 and that the T allele and T/T genotype lead to lower miR-21 levels, which are associated with T2DM. The rs13137 could change the miR-21 level, which influences insulin sensitivity and is associated with T2DM. In the current study, we also observed that another SNP in miR-21, rs1292037, was associated with T2DM, and the rs1292037C allele was a risk factor for T2DM. Although miR-21 is an important insulin metabolic molecule, few studies have paid attention to the association of rs1292037 with T2DM. In 2020, Li et al reported that the rs1292037C allele and C/C genotype in miR-21 were strongly associated with elevated susceptibility to coronary heart disease (CHD) in a Chinese Han population.33 Notably, subjects with T2DM accounted for 167 (38.30%) of 436 patients with CHD and 152 (32.27%) of 471 healthy controls in the study by Li et al. These results indicated that rs1292037 could be associated with both CHD and T2DM. Moreover, our results showed that only rs1292037-rs13137 haplotype in miR-21 was associated with T2DM. The rs1292037C-rs13137T haplotype was a risk factor for the development of T2DM and the rs1292037T-rs13137A haplotype was a protective factor of T2DM. The haplotypes results were also consistent with the single SNP results, which indicated that the association between haplotypes and T2DM could provide powerful evidence in evaluating the genetic preposition in T2DM.
miR-195 was observed to be upregulated in hyperglycaemic rat livers and to be involved the hepatic insulin signalling pathway by targeting the 3ʹUTR of INSR and repressing the expression of INSR in hepatocytes.15,16 In the current study, our results showed that in nondiabetic groups, compared with the A/G and A/A genotypes, the rs78312845G/G genotype in miR-195 was associated with lower FPG and HbA1C. FPG is mainly derived from hepatic gluconeogenesis, and elevating hepatic gluconeogenesis contributes to impaired fasting glucose.34 Activation of the insulin/INSR/AKT/GLUT insulin signalling pathway suppresses hepatic gluconeogenesis.35 Thus, the reason of the rs78312845G/G genotype associating with lower FPG and HbA1C could be rs78312845G/G genotype influence the miR-195 levels, which decreased the repression of INSR and suppressed hepatic gluconeogenesis.
One limitation of the current study is that a relatively moderate sample size may limit its statistical power; the statistical power for the effect of the rs1292037 and rs13137 in miR-21 was 0.595 and 0.528 respectively, which was relatively moderate. The other limitation is that we did not test the level of these miRNAs and INSR, IRS, and GLUT4 in plasma among our samples. Thus, we were unable to analyse the relationship between these SNPs genotypes and level of these miRNAs, INSR, IRS, and GLUT4 in plasma. A larger population and the association between SNPs genotypes and level of these miRNAs, INSR, IRS, and GLUT4 should be investigated in future studies.
Conclusions
In the current study, our data showed that rs1292037 and rs13137 in miR-21 were associated with T2DM (P=0.002 and P=0.003, respectively). The C allele of rs1292037 in miR-21 could increase the risk of T2DM (P=0.002, OR=1.17; 95% CI: 1.06–1.29), and the T allele of rs13137 in miR-21 could also be a risk factor for T2DM (P=0.003, OR=1.16; 95% CI: 1.05–1.28). Furthermore, the rs1292037C-rs13137T haplotype in miR-21 was associated with the risk of T2DM (P=0.003, OR=1.16; 95% CI:1.05–1.28), and the rs1292037T-rs13137A haplotype in miR-21 was the protection factor of T2DM (P=0.003, OR=0.86; 95% CI:0.78–0.95). Moreover, the rs78312845G/G genotype in miR-195 was associated with lower FPG and HbA1C in the nondiabetic groups (P<0.05). In the future, functional studies of rs1292037 and rs13137 of miR-21 and rs78312845 in miR-195 should be investigated.
Abbreviations
T2DM, type 2 diabetes; SNP, single nucleotide polymorphisms; NDM, nondiabetic; INSR, insulin receptor; IRS, insulin receptor substrate; GLUT4, glucose transporter 4; FPG, fasting plasma glucose; TC, total cholesterol; HDLC, high-density lipoprotein cholesterol; TG, triglycerides; LDL-C, low-density lipoprotein cholesterol; OR, odds ratio; AIC, Akaike information criterion; BIC, Bayesian information criterion.
Data Sharing Statement
The datasets used and analysed during the current study are available from Prof. Ying Yang on reasonable request (Email address: [email protected]).
Funding
This work was supported by grants from the Association Foundation Program of Yunnan Provincial Science and Technology Department and Kunming Medical University (2019FE001-092 and 202101AY070001-281), National Science Foundation of China (31660313 and 81760734), Reserve talents of young and middle-aged academic leaders in Yunnan Province (2018HB047), Special Funds for high level health talents of Yunnan Province (D-2017040), and the Clinical Medical Centre of Endocrinology and Metabolic Disease of Yunnan Province (ZX2019-02-02). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
1. IDF Diabetes Atlas 10th Edition; 2021. Available from: https://diabetesatlas.org/atlas/tenth-edition/.
2. American Diabetes A. 2. Classification and Diagnosis of Diabetes. Diabetes Care. 2017;40(Suppl1):S11–S24. doi:10.2337/dc17-S005.
3. Lizcano JM, Alessi DR. The insulin signalling pathway. Current Biol. 2002;12(7):R236–8.
4. Pessin JE, Saltiel AR. Signaling pathways in insulin action: molecular targets of insulin resistance. J Clin Invest. 2000;106(2):165–169. doi:10.1172/JCI10582
5. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297.
6. Zhang T, Lv C, Li L, et al. Plasma miR-126 is a potential biomarker for early prediction of type 2 diabetes mellitus in susceptible individuals. Biomed Res Int. 2013;2013:761617. doi:10.1155/2013/761617
7. He QF, Wang LX, Zhong JM, et al. Circulating MicroRNA-21 is downregulated in patients with metabolic syndrome. Biomed Environ Sci. 2016;29(5):385–389. doi:10.3967/bes2016.050
8. Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48(1):61–69. doi:10.1007/s00592-010-0226-0
9. Wang X, Sundquist J, Zoller B, et al. Determination of 14 circulating microRNAs in Swedes and Iraqis with and without diabetes mellitus type 2. PLoS One. 2014;9(1):e86792. doi:10.1371/journal.pone.0086792
10. Murri M, Insenser M, Fernandez-Duran E, San-Millan JL, Escobar-Morreale HF. Effects of polycystic ovary syndrome (PCOS), sex hormones, and obesity on circulating miRNA-21, miRNA-27b, miRNA-103, and miRNA-155 expression. J Clin Endocrinol Metab. 2013;98(11):E1835–44. doi:10.1210/jc.2013-2218
11. Chen YH, Heneidi S, Lee JM, et al. miRNA-93 inhibits GLUT4 and is overexpressed in adipose tissue of polycystic ovary syndrome patients and women with insulin resistance. Diabetes. 2013;62(7):2278–2286. doi:10.2337/db12-0963
12. Ortega FJ, Mercader JM, Moreno-Navarrete JM, et al. Profiling of circulating microRNAs reveals common microRNAs linked to type 2 diabetes that change with insulin sensitization. Diabetes Care. 2014;37(5):1375–1383. doi:10.2337/dc13-1847
13. Meng S, Cao JT, Zhang B, Zhou Q, Shen CX, Wang CQ. Downregulation of microRNA-126 in endothelial progenitor cells from diabetes patients, impairs their functional properties, via target gene Spred-1. J Mol Cell Cardiol. 2012;53(1):64–72. doi:10.1016/j.yjmcc.2012.04.003
14. Ghorbani S, Mahdavi R, Alipoor B, et al. Decreased serum microRNA-21 level is associated with obesity in healthy and type 2 diabetic subjects. Arch Physiol Biochem. 2018;124(4):300–305. doi:10.1080/13813455.2017.1396349
15. Herrera BM, Lockstone HE, Taylor JM, et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia. 2010;53(6):1099–1109. doi:10.1007/s00125-010-1667-2
16. Yang WM, Jeong HJ, Park SY, Lee W. Saturated fatty acid-induced miR-195 impairs insulin signaling and glycogen metabolism in HepG2 cells. FEBS Lett. 2014;588(21):3939–3946. doi:10.1016/j.febslet.2014.09.006
17. Fernandez-Twinn DS, Alfaradhi MZ, Martin-Gronert MS, et al. Downregulation of IRS-1 in adipose tissue of offspring of obese mice is programmed cell-autonomously through post-transcriptional mechanisms. Mol Metab. 2014;3(3):325–333. doi:10.1016/j.molmet.2014.01.007
18. de Almeida Faria J, Duque-Guimaraes D, Carpenter AA, Loche E, Ozanne SE. A post-weaning obesogenic diet exacerbates the detrimental effects of maternal obesity on offspring insulin signaling in adipose tissue. Sci Rep. 2017;7:44949. doi:10.1038/srep44949
19. Karolina DS, Armugam A, Tavintharan S, et al. MicroRNA 144 impairs insulin signaling by inhibiting the expression of insulin receptor substrate 1 in type 2 diabetes mellitus. PLoS One. 2011;6(8):e22839. doi:10.1371/journal.pone.0022839
20. Gaudet AD, Fonken LK, Gushchina LV, et al. miR-155 deletion in female mice prevents diet-induced obesity. Sci Rep. 2016;6:22862. doi:10.1038/srep22862
21. Ying W, Riopel M, Bandyopadhyay G, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171(2):372–384 e12. doi:10.1016/j.cell.2017.08.035
22. Ling HY, Hu B, Hu XB, et al. MiRNA-21 reverses high glucose and high insulin induced insulin resistance in 3T3-L1 adipocytes through targeting phosphatase and tensin homologue. Exp Clin Endocrinol Diabetes. 2012;120(9):553–559. doi:10.1055/s-0032-1311644
23. Shi Z, Zhao C, Guo X, et al. Differential expression of microRNAs in omental adipose tissue from gestational diabetes mellitus subjects reveals miR-222 as a regulator of ERalpha expression in estrogen-induced insulin resistance. Endocrinology. 2014;155(5):1982–1990. doi:10.1210/en.2013-2046
24. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. doi:10.1038/nature09267
25. Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91(3):827–887. doi:10.1152/physrev.00006.2010
26. Zhang Y, Li M, Bao L, Hu P. A case-control study on the relationship between miRNAs single nucleotide polymorphisms and sepsis risk. Medicine. 2019;98(33):e16744. doi:10.1097/MD.0000000000016744
27. Li Y, Li C, Yang M, et al. Association of single nucleotide polymorphisms of miRNAs involved in the GLUT4 pathway in T2DM in a Chinese population. Mol Genet Genomic Med. 2019;7(9):e907. doi:10.1002/mgg3.907
28. Buzzetti R, Tuomi T, Mauricio D, et al. Management of latent autoimmune diabetes in adults: a consensus statement from an international expert panel. Diabetes. 2020;69(10):2037–2047. doi:10.2337/dbi20-0017
29. Li Z, Zhang Z, He Z, et al. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis. Cell Res. 2009;19(4):519–523. doi:10.1038/cr.2009.33
30. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi:10.1038/sj.cr.7290272
31. Sole X, Guino E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi:10.1093/bioinformatics/btl268
32. Dupont WD, Plummer WD
33. Li H, Liu Y, Huang J, Liu Y, Zhu Y. Association of genetic variants in lncRNA GAS5/miR-21/mTOR axis with risk and prognosis of coronary artery disease among a Chinese population. J Clin Lab Anal. 2020;34(10):e23430. doi:10.1002/jcla.23430
34. Bock G, Chittilapilly E, Basu R, et al. Contribution of hepatic and extrahepatic insulin resistance to the pathogenesis of impaired fasting glucose: role of increased rates of gluconeogenesis. Diabetes. 2007;56(6):1703–1711. doi:10.2337/db06-1776
35. Das S, Choudhuri D. Dietary calcium regulates the insulin sensitivity by altering the adipokine secretion in high fat diet induced obese rats. Life Sci. 2020;250:117560. doi:10.1016/j.lfs.2020.117560
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

