Back to Journals » Cancer Management and Research » Volume 13
The Roles of Transmembrane Mucins Located on Chromosome 7q22.1 in Colorectal Cancer
Authors Almasmoum H
Received 24 December 2020
Accepted for publication 16 March 2021
Published 14 April 2021 Volume 2021:13 Pages 3271—3280
DOI https://doi.org/10.2147/CMAR.S299089
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Bilikere Dwarakanath
Hussain Almasmoum
Laboratory Medicine Department, Faculty of Applied Medical Science, Umm Al-Qura University, Makkah, 7607, Saudi Arabia
Correspondence: Hussain Almasmoum
Laboratory Medicine Department, Faculty of Applied Medical Science, Umm Al-Qura University, Al Abdeyah, Makkah, 7607, Saudi Arabia
Tel +966 125270000-ext4521
Email [email protected]
Abstract: Colorectal cancer (CRC) is one of the most common types of cancers. It is associated with a poor prognosis and high mortality. The role of mucins (MUCs) in colon tumorigenesis is unclear, but it might be significant in the progression of malignancy. Some mucins, such as MUC1 and MUC13, act as oncogenes, whereas others, such as MUC2 and MUC6, are tumor suppressors. However, there are still mucins with unidentified roles in CRC. In this review, we discuss the reported roles of mucins in CRC. Moreover, we review the capability of the mucin family to serve as a sensitive and specific histopathological marker for the early diagnosis of CRC. Lastly, the role of mucin genes clustered on chromosome 7q22 in CRC and other cancers is also discussed.
Keywords: colorectal cancer, mucins, MUC3, MUC12, MUC17, chromosome 7q22.1
Introduction
Colorectal cancer (CRC) has an annual incidence of 1.9 million cases and is the second leading cause of cancer-related death globally (935,000 deaths per year).1 The incidence of CRC is low in patients younger than 50 years old, but the rates increase thereafter, with a median age of 70 years.2,3 The lowest incidence was reported in central and southern Asia, as well as Africa. The highest incidence was reported in Europe and North America.1,4 In developed countries, the CRC incidence has stabilized or has started to decrease. This might be linked to the use of sigmoidoscopy and colonoscopy with polypectomy.5
In the Kingdom of Saudi Arabia (KSA), colorectal cancer is the most common cancer in men and the third most common in women.5 For both sexes, CRC is the most common cancer in this region, with an estimated age-standardized incidence of 88.7 cases for every 100,000 people.6 Additionally, the median ages of reported colon neoplasia among Saudi men and women are 60 and 55 years, respectively.7 These are lower than those reported (70 years old) around the world.2 The high incidence among the younger age population might be due to the relatively younger median age in this region since 71% of the population are younger than 45 years of age. Consequently, a national committee was formed by the Saudi Ministry of Health, and the panel of experts recommended that screening for CRC in KSA should be done at the age of 45 years due to the high prevalence.8,9
The treatment strategy for CRC depends on the tumor stage. For surgical intervention, polypectomy or colectomy is the first treatment option in the early stages of the disease. For advanced cases, adjuvant and neoadjuvant CRC therapies are used depending on the disease stage.10,11 Fluorouracil (5–FU) treatment is the primary therapy for CRC and some patients are resistance to this therapy and even to new chemotherapies or targeted therapies.11–15 The mechanistic basis behind the anti-cancer drug resistance in CRC might include defective drug delivery within tumor cells, impaired cellular homeostasis, and the deterioration of drug sensitivity at the molecular level. Moreover, additional mechanisms might include dysregulated programmed cell death, diminished DNA damage repair systems, and defective cell cycle checkpoints.16 This drug resistance leads to a poor prognosis and relatively lower survival rate. Half of CRC patients have a 5-year survival and this reduces with age.14,15,17 Approximately 40% of the 5-year-relative survival rate of patients with CRC.9 These rates emphasize the urgent need to develop new diagnostic approaches that are more sensitive and specific for the detection of pre-neoplastic lesions in high-risk populations to maintain better prognosis and decrease CRC-associated morbidity.
Neoplastic colon lesions develop from the stem cell niche located at the base of the colonic crypts. Colon adenoma and subsequent adenocarcinoma originate from abnormal colonic stem cell proliferation. Clinically, the diagnosis of CRC relies on the visualization of the tumor by colonoscopy or sigmoidoscopy, followed by histopathological examination of the collected tissue biopsies for confirmation.18 For earlier diagnosis of neoplastic transformation during the early stages of colonic malignancy, the detection of premalignant lesions is recommended. Aberrant crypt foci (ACF) and mucin-depleted foci (MDF) are pre-neoplastic lesions. ACF are detected via high-magnification chronoscopic colonoscopy, which enhances the detectability of ACF and flat adenomas after methylene blue staining in vivo.19,20
Only 5% of ACF were found to be premalignant, and the absence of mucin secretion by these aberrant foci suggested that there was a more specific marker for pre-neoplastic transformation of the colonic mucosa. Hence, the latter was named MDF, and they were found to be highly correlated with CRC development.21 MDF were also observed in both hereditary and sporadic CRC forms.22 Therefore, it was suggested that these pre-neoplastic lesions can be used as additional biomarkers of colon cancer.23
PubMed search engine was used to retrieve the most relevant articles in this topic24–34 and.25,35–39 The key words used to obtain the search results and the main relevant articles are summarized in flowchart (Figure 1). The related studies that mentioned in the bibliography these relevant articles were cited in this review article.
 |
Figure 1 Flowchart illustrating the search strategy in PubMed database. |
Colorectal Cancer Molecular Pathways
The sporadic of subtype of colorectal cancer accounts for ~85% and the rest include the family subtype (hereditary). CRC is traditionally classified either by chromosomal instability (CIN) or microsatellite instability (MSI). The chromosomal instability pathway accounts for approximately 80% of sporadic CRC patients.40 The first adenoma-carcinoma sequence in CRC was introduced by Fearon and Vogelstein in 1990.41 The mutation and loss of heterozygosity of several oncogenes and suppressor genes results in CIN tumors. One of the early events in the adenoma-carcinoma sequence is a mutation in the tumor suppressor gene adenomatous polyposis coli (APC). The mutation in APC leads to hyperactivation of the WNT signaling pathway in 80% of CRC patients, and other mutations in WNT components account for approximately 10%42,43 The CpG island methylator phenotype (CIMP) comprises highly frequent abnormal methylation in the CpG islands, which is reported in female and older CRC patients, and it is associated with BRAF mutations and deficiencies in mismatch repair genes.44,45
KRAS is an important member of several growth pathways, including the epidermal growth factor pathway. Constant activation of KRAS due to mutation leads to the upregulation of RAF-MEK-ERK, PI3K, and NF-KB.46 The activating mutations in the KRAS gene occur in 40% of CRC patients and is usually reported after APC mutation.47,48 Concordant mutations in KRAS and PIK3CA genes have been reported in CRC.49 Further, overactivation of the PI3K pathway inhibits the apoptosis of CRC cells.50
A late event in adenoma transition to carcinoma is the loss of heterozygosity (LOH) of chromosome 17p, which harbors the TP53 gene. Inactivation of this suppressor gene accounts for 70% of the CIN subtype of CRC patients.51 Furthermore, LOH occurs in chromosome 18, which contains the tumor suppressor genes DCC, SMAD2, and SMAD4. The latter two genes regulate the transforming growth factor (TGFB) pathway.
It is also important to highlight the molecular importance of differences between right and left colon carcinoma. In right-side colon cancer, there tends to be a high prevalence of MSI, BRAF mutations, and a high CIMP+ phenotype, and this is associated with poorer prognosis.52 For left–sided CRC, the carcinomas tend to have a high prevalence of CIN, KRAS, APC, and P53 mutations, as well as high expression of EGFR.52–54 Furthermore, chromosomal amplification has been identified in chromosomes 1q, 7, 8q, 13q, and 20q. Deletions of 1p, 4, 5q, 8p, 14q, 15q, 17p, and 18q have also been noted.55
Chromosome 7 Amplification in CRC
Chromosome 7 amplification has been reported in several studies.56–58 Several genes located on chromosome 7 have been reported to drive CRC carcinogenesis. Phosphoserine phosphatase (PSPH),59 GTF2IRD1,60 and TSA have specifically been reported to drive pathogenesis.61 Given the role of chromosome 7 amplification and its genes in CRC carcinogenesis, MUC3, MUC12, and MUC17 mucins, encoded by the same chromosome, have been implicated in CRC pathogenesis.
Mucins
Mucins are glycosylated proteins that are synthesized and expressed by a variety of tissues, including the colon.62 Mucins can be categorized into three categories as follows: (i) membrane-bound/transmembrane mucins, (ii) secreted (gel-forming), and (iii) soluble (non-gel-forming) mucins.62 Mucins have a surface protective role for epithelial cells via the entrapment of pathogens.63 They are also involved in cell signaling pathways.63 Therefore, they have a fundamental role in cellular functions, mainly at the surface of epithelial cells (Table 1). The molecular identification of mucins has revealed more than 20 mucinous proteins (MUC). These include membrane mucins, such as MUC1, MUC3A, MUC3B, MUC4, MUC12, MUC13, MUC15, MUC16, MUC17, and MUC2, and secreted mucins, such as MUC2, MUC5AC, MUC5B, MUC6, and MUC19. Further, MUC7, MUC8, MUC9, and MUC20 are soluble mucins.62
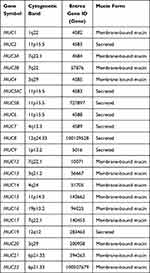 |
Table 1 Mucin Genes and Cytogenetic Localization |
The primary function of mucins is to protect the surface of epithelial tissues. However, several mucins have been found to be pathologically expressed and involved in the tumorigenesis of various solid tumors. Additionally, published studies on MUCs in CRC are limited and only include certain members of the mucin family. MUC1 is expressed by both neoplastic and normal colonic tissues. Its expression is significantly higher in malignant lesions and is associated with a worse prognosis.64 However, no correlation was observed between MUC1 expression and MSI CRC cases.65 Additionally, the upregulation of MUC1 with β-catenin in gastric and colon neoplasia is considered a predictor of worse prognosis.66,67 However, the proposed underlying pathogenic mechanisms associated with MUC1 are conflicting. Several studies have suggested that MUC1 induces cell proliferation and invasion by binding to β-catenin, thus inducing nuclear translocation of the latter.68 Other studies have proposed that MUC1 suppresses cellular proliferation by preventing β-catenin nuclear localization.69 More studies are needed to illustrate the precise role of MUC1 in CRC.
Similarly, contradictory results related to the role of MUC4 in colon tumorigenesis have been reported. Overexpression of MUC4 has been reported in tissues obtained from colon malignancies and is associated with worse prognosis.70 However, others also reported the loss of MUC4 in tissue samples obtained from CRC.71 Another membrane mucin, MUC13, was also found to be upregulated in primary and metastatic CRC.72,73 Its overexpression has recently been linked to the development of resistance to chemotherapy in patients diagnosed with CRC.74
The oncogenic role of MUC15 in CRC has been reported, in which overexpression was reported in CRC tissues compared to levels in the non-tumor tissue of the same cases.75 The effects of MUC15 in vitro in CRC cell lines have been studied, in which MUC15 was determined to increase proliferation and cell motility.75 MUC16 (known as CA125) is the best biomarker for monitoring the progression of ovarian cancer and also can be used to monitor progression and predict lymph node metastasis in CRC cases.76,77
Alternatively, the overexpression of other mucins has been shown to be favorable for the prognosis of CRC. MUC2 is the most commonly secreted mucin in the intestines,71 and mice deficient in the MUC2 gene (Muc2−/−) spontaneously develop colon cancer.78,79 Furthermore, MUC2 is downregulated in CRC tissues obtained from humans, which suggests its tumor suppressor role.80,81 Therefore, an increase in MUC2 can protected and/or enhance the outcome of CRC. Correspondingly, an increase in the expression of both MUC5AC and MUC6 also contribute to a better outcome for CRC.34,49,71,82
Regarding soluble forms, aberrant glycopeptides of MUC7 were not detected in CRC cases.83 Nevertheless, MUC7 expression is related to the recurrence of bladder cancer.84 However, other mucins like MUC8, MUC9, and MUC20 have not been studied in CRC. The serum level of MUC9 (known as OVGP1) increases with ovarian cancer stages.85 In addition, the overexpression of MUC20 was reported in endometrial cancer, and it is associated with tumorigenesis and poor survival.86
Notably, genes encoding secreted mucins (MUC2, MUC5AC, MUC5B, MUC6, and MUC19) are all clustered-on chromosome 11 and are reportedly co-expressed.87 Furthermore, recent studies have also explored the expression of secreted mucins located on chromosome 11 in a large series of CRC,87 demonstrating that the overexpression of MUC5AC, MUC5B, and MUC6 is associated with serrated types of colonic glandular neoplasia. Serrated CRC is associated with DNA hypermethylation, MSI, and BRAF somatic mutations.87 Additionally, mucin genes contain numerous transcription factor sites, such as those for Sp1, SP3, AP-1, NFκ-B, and CDX2.68 The loss of CDX2 expression has been reported in several studies, due to mutation or microsatellite repeats within CDX2 or through epigenetic silencing.88,89 CDX2 expression was found in only 5% of CRC patients, and in another study, it was determined to be completely lost.87,90 Downregulation of this transcription factor upregulates the expression of MUC2, MUC5AC, and MUC6. Furthermore, epigenetic regulation of genes expressing mucins in chromosome 11p15 has been explored by Vincent et al in several epithelial cancer cell lines, which include esophageal, pancreatic, gastric, and colon.91 They showed that MUC2 and MUCB genes are mainly affected by epigenetic changes in which MUC2 expression is controlled by the repressive histone code and MUC5B methylation at CpG sites controls expression. However, one study has reported that MUC6 expression is not affected by epigenetic changes, and the MUC5AC epigenetic mechanism has not been fully understood.91 Regarding MUC5AC expression, hypomethylation was reported in MSI CRC cases and it is associated with poor differentiation and BRAF mutations.92 Furthermore, expression of MUC2 was reported to be associated with poorly differentiated cases and mucinous carcinoma.92
There are several therapeutical approaches for CRC treatment have been significantly improved the last decades. Drug combination has been used to enhance the effect of chemotherapy such as FOLFOX (5-FU +Oxaliplatin), however most of these approaches are failed to prevent the progression of CRC.93 Therefore, the novel therapeutical approaches are urgent required. Mucins are the essential targets for prevent progression and metastasis of various type of cancers include CRC. Table 2 summarized some of clinical trial and anticancer therapeuticaltargeted mucins by using either vaccine or immunotherapy.94–96
 |
Table 2 Therapeutic Targets of Mucins in Several Cancer Types |
Transmembrane Mucins Located on Chromosome 7q22.1
The transmembrane mucins MUC3A/B, MUC12, and MUC17 have short amino-terminal domains in the extracellular region, followed by long (> 4000 amino acids) heavily O-glycosylated tandem repeat domains. This structure is followed by two Cys-rich motifs (CRD1 and CRD2), proximal to the membrane, and both have a similar structure to the epidermal growth factor (EGF) domain.38 In addition, these two Cys-rich motifs are separated by a Linker-SEA (L) domain, which contains a sea urchin sperm protein, enterokinase, and agrin.25 Furthermore, these two Cys-rich motifs are followed by a small cytoplasmic domain. Notably, the transmembrane mucin genes MUC3A, MUC3B, MUC12, and MUC17 are clustered on chromosome 7q22.
MUC3
The expression of MUC3 has been observed in normal colon and colon malignancies.26,32 Several clinical studies have suggested an association between poor prognosis and MUC3 expression. Increased expression of MUC3 in pancreatic intraepithelial neoplasia is associated with the progression of neoplasia and is negatively correlated with differentiation.30 In pancreatic ductal cell adenocarcinoma, the protein expression of MUC3 distinguishes the malignant tissue from normal tissue.24 Furthermore, among 1447 cases of invasive breast carcinoma, the overexpression of MUC3 was found to be correlated with local recurrence and the lymph node stage, and this suggested the role of MUC3 as a prognostic marker.28 Moreover, MUC3 is associated with gastric cancer progression.29 The upregulation of MUC3 was also noted in clinical CRC samples in 84% of the cases. Among these, cytoplasmic localization and membrane localization were found in 91% and 38% of the cases, respectively.26 Others, however, have shown the opposite findings wherein decreased MUC3 protein expression was reported in CRC.31,32,97
MUC12
MUC12 has a structure similar to MUC3 and MUC17.98 Few studies have evaluated MUC12 in CRC and other cancer types. Matsuyama et al studied the expression of MUC12 mRNA in CRC. The expression of MUC12 was lower in cancer than in normal colonic tissue, and low expression was found to be associated with a poorer survival rate.34 Furthermore, CRC tissues have lower expression of MUC12 than non-tumor tissues.99 MUC12 was also determined to be expressed more in renal cell carcinoma than in the normal kidney and when knocked down. Growth and migration are decreased.33
MUC17
MUC17 harbors two EGF-like domains and is genetically similar to rodent MUC339 Several studies have examined the recombinant mouse Muc3-CRD1-L-CRD2.25,27 This recombinant protein increases cell motility and migration, inhibits apoptosis in vitro, and promotes wound healing in vivo.27 MUC17 showed a similar effect on a CRC cell line.25 In addition, clinical studies on MUC17 expression have been conducted in several cancers. Aberrant expression of MUC17 protein in pancreatic ductal adenocarcinomas has been reported.37,38 In addition, the downregulation of MUC17 protein in breast cancer is associated with a longer survival rate in patients treated with chemotherapy.35 Furthermore, high expression of MUC17 in gastric carcinoma is associated with better prognosis.100 The hypermethylation of MUC17 in gastric cancer was determined to be associated with cancer development in Helicobacter pylori infection.36 Moreover, sessile serrated adenoma/polyps (SSA/P) show the upregulation of MUC17.101 The SSA/P pathways are associated with a high frequency of MSI and CIMP, as well as with BRAF mutations.52 The overexpression of MUC17 protein in the CRC SSA/P pathway, with its distinct molecular features, suggest the possibility to further evaluate of MUC17 as a biomarker for the MSI and CIMP pathway.
Conclusion
The association among MUC17 in CRC, BRAF mutations, MSI, and CIMP needs further evaluation. The expression patterns of MUC3, MUC12, and MUC17, as well as their associations with histopathological features and clinical outcomes, have not been studied in CRC. These three mucins require further investigations involving larger clinical sample sizes. The biological activities of these three mucins in CRC also need to be explored in vitro and in vivo.
Abbreviations
ACF, aberrant crypt foci; AOM, azoxymethane; CIN, chromosomal instability; CRC, colorectal cancer; EGF, epidermal growth factor; KSA, Kingdom of Saudi Arabia; MSI, microsatellite instability; MDF, mucin-depleted foci; MUC, mucinous proteins; PSPH, phosphoserine phosphatase.
Acknowledgment
This work was supported by Deanship of Scientific Research at Umm Al-Qura University.
Funding
This project was funded by the DEANSHIP OF SCIENTIFIC RESEARCH AT UMM AL-QURA UNIVERSITY; Project (18-MED-1-01-0012). The funding organization had no role in the study design, data collection, analysis, interpretation, or manuscript writing.
Disclosure
The author reports no conflicts of interest in this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. ACS. 2020.
2. Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012;62(4):220–241. doi:10.3322/caac.21149
3. Keum N, Giovannucci E. Global burden of colorectal cancer. Nat Rev Gastroenterol Hepatol. 2019;16(12):713–732. doi:10.1038/s41575-019-0189-8
4. Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–378. doi:10.3322/caac.20038
5. Stock C, Pulte D, Haug U, Brenner H. Subsite-specific colorectal cancer risk in the colorectal endoscopy era. Gastrointest Endosc. 2012;75(3):621–630. doi:10.1016/j.gie.2011.10.025
6. Almatroudi A. The incidence rate of colorectal cancer in Saudi Arabia: an observational descriptive epidemiological analysis. Int J Gen Med. 2020;13:977–990. doi:10.2147/IJGM.S277272
7. Zubaidi AM, AlSubaie NM, AlHumaid AA, Shaik SA, AlKhayal KA, AlObeed OA. Public awareness of colorectal cancer in Saudi Arabia: a survey of 1070 participants in Riyadh. Saudi J Gastroenterol. 2015;21(2):78. doi:10.4103/1319-3767.153819
8. Mosli MH, Al-Ahwal MS. Colorectal cancer in the Kingdom of Saudi Arabia: need for screening. APJCP. 2012;13(8):3809–3813. doi:10.7314/apjcp.2012.13.8.3809
9. Alsanea N, Abduljabbar AS, Alhomoud S, Ashari LH, Hibbert D, Bazarbashi SJ. Colorectal cancer in Saudi Arabia: incidence, survival, demographics and implications for national policies. Ann Saudi Med. 2015;35(3):196–202. doi:10.5144/0256-4947.2015.196
10. Schmoll H, Van Cutsem E, Stein A, et al. ESMO consensus guidelines for management of patients with colon and rectal cancer. a personalized approach to clinical decision making. Ann Oncol. 2012;23(10):2479–2516. doi:10.1093/annonc/mds236
11. Hammond WA, Swaika A, Mody KJT. Pharmacologic resistance in colorectal cancer: a review. Ther Adv Med Oncol. 2016;8(1):57–84. doi:10.1177/1758834015614530
12. Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–117. doi:10.3322/caac.21220
13. Chua W, Kho PS, Moore MM, Charles KA, Clarke SJ. Clinical, laboratory and molecular factors predicting chemotherapy efficacy and toxicity in colorectal cancer. Crit Rev Oncol Hematol. 2011;79(3):224–250. doi:10.1016/j.critrevonc.2010.07.012
14. Sankaranarayanan R, Swaminathan R, Brenner H, et al. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol. 2010;11(2):165–173. doi:10.1016/S1470-2045(09)70335-3
15. Brenner H, Bouvier AM, Foschi R, et al. Progress in colorectal cancer survival in Europe from the late 1980s to the early 21st century: the EUROCARE study. Int J Cancer. 2012;131(7):1649–1658. doi:10.1002/ijc.26192
16. Boussios S, Ozturk MA, Moschetta M, et al. The developing story of predictive biomarkers in colorectal cancer. J Pers Med. 2019;9(1):12. doi:10.3390/jpm9010012
17. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet (London, England). 2014;383(9927):1490–1502. doi:10.1016/S0140-6736(13)61649-9
18. Zauber AG, Winawer SJ, O’Brien MJ, et al. Colonoscopic polypectomy and long-term prevention of colorectal-cancer deaths. N Engl J Med. 2012;366(8):687–696. doi:10.1056/NEJMoa1100370
19. Regueiro CR; Committee AGAFT. AGA future trends committee report: colorectal cancer: a qualitative review of emerging screening and diagnostic technologies. Gastroenterology. 2005;129(3):1083–1103. doi:10.1053/j.gastro.2005.06.012
20. Hurlstone DP, Fujii T. Practical uses of chromoendoscopy and magnification at colonoscopy. Gastrointest Endosc Clin N Am. 2005;15(4):687–702. doi:10.1016/j.giec.2005.08.014
21. Caderni G, Femia AP, Giannini A, et al. Identification of mucin-depleted foci in the unsectioned colon of azoxymethane-treated rats: correlation with carcinogenesis. Cancer Res. 2003;63(10):2388–2392.
22. Femia AP, Giannini A, Fazi M, et al. Identification of mucin depleted foci in the human colon. Cancer Prev Res (Phila). 2008;1(7):562–567. doi:10.1158/1940-6207.CAPR-08-0125
23. Femia AP, Dolara P, Caderni G. Mucin-depleted foci (MDF) in the colon of rats treated with azoxymethane (AOM) are useful biomarkers for colon carcinogenesis. Carcinogenesis. 2004;25(2):277–281. doi:10.1093/carcin/bgh005
24. Sierzega M, Młynarski D, Tomaszewska R, Kulig J. Semiquantitative immunohistochemistry for mucin (MUC1, MUC2, MUC3, MUC4, MUC5AC, and MUC6) profiling of pancreatic ductal cell adenocarcinoma improves diagnostic and prognostic performance. Histopathology. 2016;69(4):582–591. doi:10.1111/his.12994
25. Ho SB, Luu Y, Shekels LL, et al. Activity of recombinant cysteine-rich domain proteins derived from the membrane-bound MUC17/Muc3 family mucins. Biochim Biophys Acta. 2010;1800(7):629–638. doi:10.1016/j.bbagen.2010.03.010
26. Duncan TJ, Watson NFS, Al-Attar AH, Scholefield JH, Durrant LG. The role of MUC1 and MUC3 in the biology and prognosis of colorectal cancer. World J Surg Oncol. 2007;5(1):31. doi:10.1186/1477-7819-5-31
27. Ho SB, Dvorak LA, Moor RE, et al. Cysteine-rich domains of muc3 intestinal mucin promote cell migration, inhibit apoptosis, and accelerate wound healing. Gastroenterology. 2006;131(5):1501–1517. doi:10.1053/j.gastro.2006.09.006
28. Rakha EA, Boyce RW, Abd El-Rehim D, et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Modern Pathol. 2005;18(10):1295–1304. doi:10.1038/modpathol.3800445
29. Wang RQ, Fang DC. Alterations of MUC1 and MUC3 expression in gastric carcinoma: relevance to patient clinicopathological features. J Clin Pathol. 2003;56(5):378–384. doi:10.1136/jcp.56.5.378
30. Park HU, Kim JW, Kim GE, et al. Aberrant expression of MUC3 and MUC4 membrane-associated mucins and sialyl Le(x) antigen in pancreatic intraepithelial neoplasia. Pancreas. 2003;26(3):e48–54. doi:10.1097/00006676-200304000-00022
31. Cao Y, Schlag PM, Karsten U. Immunodetection of epithelial mucin (MUC1, MUC3) and mucin-associated glycotopes (TF, Tn, and sialosyl-Tn) in benign and malignant lesions of colonic epithelium: apolar localization corresponds to malignant transformation. Virchows Archiv. 1997;431(3):159–166. doi:10.1007/s004280050083
32. Chang S-K, Dohrman AF, Basbaum CB, et al. Localization of mucin (MUC2 and MUC3) messenger RNA and peptide expression in human normal intestine and colon cancer. Gastroenterology. 1994;107(1):28–36. doi:10.1016/0016-5085(94)90057-4
33. Gao S-L, Yin R, Zhang L-F, et al. The oncogenic role of MUC12 in RCC progression depends on c-Jun/TGF-β signalling. J Cell Mol Med. 2020;24(15):8789–8802. doi:10.1111/jcmm.15515
34. Matsuyama T, Ishikawa T, Mogushi K, et al. MUC12 mRNA expression is an independent marker of prognosis in stage II and stage III colorectal cancer. Int J Cancer. 2010;127(10):2292–2299. doi:10.1002/ijc.25256
35. Al Amri WS, Allinson LM, Baxter DE, et al. Genomic and expression analyses define MUC17 and PCNX1 as predictors of chemotherapy response in breast cancer. Mol Cancer Ther. 2020;19(3):945–955. doi:10.1158/1535-7163.MCT-19-0940
36. Lin S, Zhang Y, Hu Y, et al. Epigenetic downregulation of MUC17 by H. pylori infection facilitates NF-κB-mediated expression of CEACAM1-3S in human gastric cancer. Gastric Cancer. 2019;22(5):941–954. doi:10.1007/s10120-019-00932-0
37. Kitamoto S, Yokoyama S, Higashi M, et al. Expression of MUC17 is regulated by HIF1α-mediated hypoxic responses and requires a methylation-free hypoxia responsible element in pancreatic cancer. PLoS One. 2012;7(9):e44108–e44108. doi:10.1371/journal.pone.0044108
38. Moniaux N, Junker WM, Singh AP, Jones AM, Batra SK. Characterization of human mucin MUC17. Complete coding sequence and organization. J Biol Chem. 2006;281(33):23676–23685. doi:10.1074/jbc.M600302200
39. Gum JR, Crawley SC, Hicks JW, Szymkowski DE, Kim YS. MUC17, a novel membrane-tethered mucin. Biochem Biophys Res Commun. 2002;291(3):466–475. doi:10.1006/bbrc.2002.6475
40. Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med. 2009;361(25):2449–2460. doi:10.1056/NEJMra0804588
41. Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi:10.1016/0092-8674(90)90186-i
42. Morin PJ, Sparks AB, Korinek V, et al. Activation of β-catenin-Tcf signaling in colon cancer by mutations in β-catenin or APC. Science (New York, N.Y.). 1997;275(5307):1787–1790. doi:10.1126/science.275.5307.1787
43. Silva A-L, Dawson SN, Arends MJ, et al. Boosting Wnt activity during colorectal cancer progression through selective hypermethylation of Wnt signaling antagonists. BMC Cancer. 2014;14(1):1–10.
44. Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi:10.1056/NEJMoa1500596
45. Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi:10.1038/ng1834
46. Pruitt K, Der CJ. Ras and Rho regulation of the cell cycle and oncogenesis. Cancer Lett. 2001;171(1):1–10. doi:10.1016/S0304-3835(01)00528-6
47. Santini D, Loupakis F, Vincenzi B, et al. High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist. 2008;13(12):1270–1275. doi:10.1634/theoncologist.2008-0181
48. Tsuchida N, Ohtsubo E, Ryder TJS. Nucleotide sequence of the oncogene encoding the p21 transforming protein of Kirsten murine sarcoma virus. Science (New York, N.Y.). 1982;217(4563):937–939. doi:10.1126/science.6287573
49. De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–762. doi:10.1016/S1470-2045(10)70130-3
50. Benvenuti S, Frattini M, Arena S, et al. PIK3CA cancer mutations display gender and tissue specificity patterns. Human Mutation. 2008;29(2):284–288. doi:10.1002/humu.20648
51. Nature CGANJ. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330.
52. Zarkavelis G, Boussios S, Papadaki A, Katsanos KH, Christodoulou DK, Pentheroudakis G. Current and future biomarkers in colorectal cancer. Ann Gastroenterol. 2017;30(6):613–621. doi:10.20524/aog.2017.0191
53. Schell M, Yang M, Teer J, et al. A multigene mutation classification of 468 colorectal cancers reveals a prognostic role for APC. Nat Commun. 2016;7:11743. doi:10.1038/ncomms11743
54. Kaz AM, Wong C-J, Dzieciatkowski S, Luo Y, Schoen RE, Grady WM. Patterns of DNA methylation in the normal colon vary by anatomical location, gender, and age. Epigenetics. 2014;9(4):492–502. doi:10.4161/epi.27650
55. Xu J-F, Kang Q, Ma X-Y, et al. A novel method to detect early colorectal cancer based on chromosome copy number variation in plasma. Cell Physiol Biochem. 2018;45(4):1444–1454. doi:10.1159/000487571
56. Herbergs J, Arends JW, Bongers EM, Ramaekers FC, Hopman AH. Clonal origin of trisomy for chromosome 7 in the epithelial compartment of colon neoplasia. Genes Chromosomes Cancer. 1996;16(2):106–112. doi:10.1002/(SICI)1098-2264(199606)16:2<106::AID-GCC4>3.0.CO;2-3
57. Uchi R, Takahashi Y, Niida A, et al. Integrated multiregional analysis proposing a new model of colorectal cancer evolution. PLoS Genet. 2016;12(2):e1005778. doi:10.1371/journal.pgen.1005778
58. Zarzour P, Boelen L, Luciani F, et al. Single nucleotide polymorphism array profiling identifies distinct chromosomal aberration patterns across colorectal adenomas and carcinomas. Genes Chromosomes Cancer. 2015;54(5):303–314. doi:10.1002/gcc.22243
59. Sato K, Masuda T, Hu Q, et al. Phosphoserine phosphatase is a novel prognostic biomarker on chromosome 7 in colorectal cancer. Anticancer Res. 2017;37(5):2365–2371. doi:10.21873/anticanres.11574
60. Nambara S, Masuda T, Kobayashi Y, et al. GTF2IRD1 on chromosome 7 is a novel oncogene regulating the tumor-suppressor gene TGFβR2 in colorectal cancer. Cancer Sci. 2020;111(2):343–355. doi:10.1111/cas.14248
61. Buishand FO, Cardin E, Hu Y, Ried T. Trichostatin A preferentially reverses the upregulation of gene-expression levels induced by gain of chromosome 7 in colorectal cancer cell lines. Genes Chromosomes Cancer. 2018;57(1):35–41. doi:10.1002/gcc.22505
62. Andrianifahanana M, Moniaux N, Batra SK. Regulation of mucin expression: mechanistic aspects and implications for cancer and inflammatory diseases. Biochim Biophys Acta. 2006;1765(2):189–222. doi:10.1016/j.bbcan.2006.01.002
63. Kaur S, Kumar S, Momi N, Sasson AR, Batra SK. Mucins in pancreatic cancer and its microenvironment. Nat Rev Gastroenterol Hepatol. 2013;10(10):607–620. doi:10.1038/nrgastro.2013.120
64. Kesari MV, Gaopande VL, Joshi AR, Babanagare SV, Gogate BP, Khadilkar AV. Immunohistochemical study of MUC1, MUC2 and MUC5AC in colorectal carcinoma and review of literature. Indian J Gastroenterol. 2015;34(1):63–67. doi:10.1007/s12664-015-0534-y
65. Biemer-Huttmann AE, Walsh MD, McGuckin MA, et al. Mucin core protein expression in colorectal cancers with high levels of microsatellite instability indicates a novel pathway of morphogenesis. Clin Cancer Res. 2000;6(5):1909–1916.
66. Retterspitz MF, Monig SP, Schreckenberg S, et al. Expression of {beta}-catenin, MUC1 and c-met in diffuse-type gastric carcinomas: correlations with tumour progression and prognosis. Anticancer Res. 2010;30(11):4635–4641.
67. Baldus SE, Monig SP, Huxel S, et al. MUC1 and nuclear beta-catenin are coexpressed at the invasion front of colorectal carcinomas and are both correlated with tumor prognosis. Clin Cancer Res. 2004;10(8):2790–2796. doi:10.1158/1078-0432.CCR-03-0163
68. Wang Z, Sun J, Hu X, Huang S. Interference of mucin 1 inhibits progression of colon carcinoma by repression of Wnt/beta-catenin signaling. DNA Cell Biol. 2014;33(3):162–170. doi:10.1089/dna.2013.2274
69. Lillehoj EP, Lu W, Kiser T, Goldblum SE, Kim KC. MUC1 inhibits cell proliferation by a beta-catenin-dependent mechanism. Biochim Biophys Acta. 2007;1773(7):1028–1038. doi:10.1016/j.bbamcr.2007.04.009
70. Shanmugam C, Jhala NC, Katkoori VR, et al. Prognostic value of mucin 4 expression in colorectal adenocarcinomas. Cancer. 2010;116(15):3577–3586. doi:10.1002/cncr.25095
71. Biemer-Huttmann AE, Walsh MD, McGuckin MA, et al. Immunohistochemical staining patterns of MUC1, MUC2, MUC4, and MUC5AC mucins in hyperplastic polyps, serrated adenomas, and traditional adenomas of the colorectum. J Histochem Cytochem. 1999;47(8):1039–1048. doi:10.1177/002215549904700808
72. Walsh MD, Young JP, Leggett BA, Williams SH, Jass JR, McGuckin MA. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Hum Pathol. 2007;38(6):883–892. doi:10.1016/j.humpath.2006.11.020
73. Gupta BK, Maher DM, Ebeling MC, et al. Increased expression and aberrant localization of mucin 13 in metastatic colon cancer. J Histochem Cytochem. 2012;60(11):822–831. doi:10.1369/0022155412460678
74. Sheng YH, He Y, Hasnain SZ, et al. MUC13 protects colorectal cancer cells from death by activating the NF-kappaB pathway and is a potential therapeutic target. Oncogene. 2017;36(5):700–713. doi:10.1038/onc.2016.241
75. Huang J, Che M-I, Huang Y-T, et al. Overexpression of MUC15 activates extracellular signal-regulated kinase 1/2 and promotes the oncogenic potential of human colon cancer cells. Carcinogenesis. 2009;30(8):1452–1458. doi:10.1093/carcin/bgp137
76. Duffy M, Bonfrer J, Kulpa J, et al. CA125 in ovarian cancer: European Group on Tumor Markers guidelines for clinical use. Int J Gynecol Cancer. 2005;15(5):679–691. doi:10.1111/j.1525-1438.2005.00130.x
77. Gao Y, Wang J, Zhou Y, Sheng S, Qian SY, Huo X. Evaluation of serum CEA, CA19-9, CA72-4, CA125 and ferritin as diagnostic markers and factors of clinical parameters for colorectal cancer. Sci Rep. 2018;8(1):2732. doi:10.1038/s41598-018-21048-y
78. Velcich A, Yang W, Heyer J, et al. Colorectal cancer in mice genetically deficient in the mucin Muc2. J Science. 2002;295(5560):1726–1729.
79. Van der Sluis M, De Koning BA, De Bruijn AC, et al. Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology. 2006;131(1):117–129. doi:10.1053/j.gastro.2006.04.020
80. Imai Y, Yamagishi H, Fukuda K, Ono Y, Inoue T, Ueda Y. Differential mucin phenotypes and their significance in a variation of colorectal carcinoma. World J Gastroenterol. 2013;19(25):3957–3968. doi:10.3748/wjg.v19.i25.3957
81. Ogata S, Uehara H, Chen A, Itzkowitz SH. Mucin gene expression in colonic tissues and cell lines. Cancer Res. 1992;52(21):5971–5978.
82. Betge J, Schneider NI, Harbaum L, et al. MUC1, MUC2, MUC5AC, and MUC6 in colorectal cancer: expression profiles and clinical significance. Virchows Arch. 2016;469(3):255–265. doi:10.1007/s00428-016-1970-5
83. Pedersen JW, Blixt O, Bennett EP, et al. Seromic profiling of colorectal cancer patients with novel glycopeptide microarray. Int J Cancer. 2011;128(8):1860–1871. doi:10.1002/ijc.25778
84. Han Y, Zheng Q, Tian Y, Ji Z, Ye H. Identification of a nine-gene panel as a prognostic indicator for recurrence with muscle-invasive bladder cancer. J Surg Oncol. 2019;119(8):1145–1154. doi:10.1002/jso.25446
85. Maines-Bandiera S, Woo MM, Borugian M, et al. Oviductal glycoprotein (OVGP1, MUC9): a differentiation-based mucin present in serum of women with ovarian cancer. Int J Gynecol Cancer. 2010;20(1):16–22. doi:10.1111/IGC.0b013e3181bcc96d
86. Zheng F, Yu H, Lu J. High expression of MUC20 drives tumorigenesis and predicts poor survival in endometrial cancer. J Cell Biochem. 2019.
87. Walsh MD, Clendenning M, Williamson E, et al. Expression of MUC2, MUC5AC, MUC5B, and MUC6 mucins in colorectal cancers and their association with the CpG island methylator phenotype. Modern Pathol. 2013;26(12):1642–1656. doi:10.1038/modpathol.2013.101
88. Wicking C, Simms LA, Evans T, et al. CDX2, a human homologue of Drosophila caudal, is mutated in both alleles in a replication error positive colorectal cancer. Oncogene. 1998;17(5):657–659. doi:10.1038/sj.onc.1201971
89. Kawai H, Tomii K, Toyooka S, et al. Promoter methylation downregulates CDX2 expression in colorectal carcinomas. Oncol Rep. 2005;13(3):547–551.
90. Mochizuka A, Uehara T, Nakamura T, Kobayashi Y, Ota H. Hyperplastic polyps and sessile serrated ‘adenomas’ of the colon and rectum display gastric pyloric differentiation. Histochem Cell Biol. 2007;128(5):445–455. doi:10.1007/s00418-007-0326-2
91. Vincent A, Perrais M, Desseyn JL, Aubert JP, Pigny P, Van Seuningen I. Epigenetic regulation (DNA methylation, histone modifications) of the 11p15 mucin genes (MUC2, MUC5AC, MUC5B, MUC6) in epithelial cancer cells. Oncogene. 2007;26(45):6566–6576. doi:10.1038/sj.onc.1210479
92. Renaud F, Vincent A, Mariette C, et al. MUC5AC hypomethylation is a predictor of microsatellite instability independently of clinical factors associated with colorectal cancer. Int J Cancer. 2015;136(12):2811–2821. doi:10.1002/ijc.29342
93. Xie Y-H, Chen Y-X, Fang J-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther. 2020;5(1):22.
94. Kufe DW. Mucins in cancer: function, prognosis and therapy. Nat Rev Cancer. 2009;9(12):874–885. doi:10.1038/nrc2761
95. Chen CH, Wu YJ, Chen JJ. Gold nanotheranostics: photothermal therapy and imaging of Mucin 7 conjugated antibody nanoparticles for urothelial cancer. Biomed Res Int. 2015;2015:813632.
96. Mohammadi M, Nejatollahi F, Ghasemi Y, Faraji SN. Anti-metastatic and anti-invasion effects of a specific anti-MUC18 scFv antibody on breast cancer cells. Appl Biochem Biotechnol. 2017;181(1):379–390. doi:10.1007/s12010-016-2218-1
97. Berretta M, Alessandrini L, De Divitiis C, et al. Serum and tissue markers in colorectal cancer: state of art. Crit Rev Oncol Hematol. 2017;111:103–116. doi:10.1016/j.critrevonc.2017.01.007
98. Desseyn JL, Tetaert D, Gouyer V. Architecture of the large membrane-bound mucins. Gene. 2008;410(2):215–222. doi:10.1016/j.gene.2007.12.014
99. Packer LM, Williams SJ, Callaghan S, Gotley DC, McGuckin MA. Expression of the cell surface mucin gene family in adenocarcinomas. Int J Oncol. 2004;25(4):1119–1126.
100. Yang B, Wu A, Hu Y, et al. Mucin 17 inhibits the progression of human gastric cancer by limiting inflammatory responses through a MYH9-p53-RhoA regulatory feedback loop. J Exp Clin Cancer Res. 2019;38(1):283. doi:10.1186/s13046-019-1279-8
101. Krishn SR, Kaur S, Sheinin YM, et al. Mucins and associated O-glycans based immunoprofile for stratification of colorectal polyps: clinical implication for improved colon surveillance. Oncotarget. 2017;8(4):7025–7038. doi:10.18632/oncotarget.12347
102. Bitler BG, Menzl I, Huerta CL, et al. Intracellular MUC1 peptides inhibit cancer progression. Clin Cancer Res. 2009;15(1):100–109. doi:10.1158/1078-0432.CCR-08-1745
103. Rubinstein DB, Karmely M, Pichinuk E, et al. The MUC1 oncoprotein as a functional target: immunotoxin binding to alpha/beta junction mediates cell killing. Int J Cancer. 2009;124(1):46–54. doi:10.1002/ijc.23910
104. Raina D, Ahmad R, Joshi MD, et al. Direct targeting of the mucin 1 oncoprotein blocks survival and tumorigenicity of human breast carcinoma cells. Cancer Res. 2009;69(12):5133–5141. doi:10.1158/0008-5472.CAN-09-0854
105. Quoix E, Lena H, Losonczy G, et al. TG4010 immunotherapy and first-line chemotherapy for advanced non-small-cell lung cancer (TIME): results from the phase 2b part of a randomised, double-blind, placebo-controlled, phase 2b/3 trial. Lancet Oncol. 2016;17(2):212–223. doi:10.1016/S1470-2045(15)00483-0
106. Wrona A. Role of immunotherapy in stage III nonsmall cell lung cancer. Curr Opin Oncol. 2019;31(1):18–23. doi:10.1097/CCO.0000000000000493
107. Sangha R, North S. L-BLP25: a MUC1-targeted peptide vaccine therapy in prostate cancer. Expert Opin Biol Ther. 2007;7(11):1723–1730. doi:10.1517/14712598.7.11.1723
108. Gatti-Mays ME, Redman JM, Donahue RN, et al. A phase I trial using a multitargeted recombinant adenovirus 5 (CEA/MUC1/brachyury)-based immunotherapy vaccine regimen in patients with advanced cancer. Oncologist. 2020;25(6):479–e899. doi:10.1634/theoncologist.2019-0608
109. Chen S, Lin Y, Zhong S, et al. 33O - Anti-MUC1 CAR-T cells combined with PD-1 knockout engineered T cells for patients with non-small cell lung cancer (NSCLC): a pilot study. Ann Oncol. 2018;29:x11. doi:10.1093/annonc/mdy485.002
110. Buckman R, De Angelis C, Shaw P, et al. Intraperitoneal therapy of malignant ascites associated with carcinoma of ovary and breast using radioiodinated monoclonal antibody 2G3. Gynecol Oncol. 1992;47(1):102–109. doi:10.1016/0090-8258(92)90084-V
111. Maraveyas A, Stafford N, Rowlinson-Busza G, Stewart JS, Epenetos AA. Pharmacokinetics, biodistribution, and dosimetry of specific and control radiolabeled monoclonal antibodies in patients with primary head and neck squamous cell carcinoma. Cancer Res. 1995;55(5):1060–1069.
112. Nishii Y, Yamaguchi M, Kimura Y, et al. A newly developed anti-Mucin 13 monoclonal antibody targets pancreatic ductal adenocarcinoma cells. Int J Oncol. 2015;46(4):1781–1787. doi:10.3892/ijo.2015.2880
113. Aithal A, Rauth S, Kshirsagar P, et al. MUC16 as a novel target for cancer therapy. Expert Opin Ther Targets. 2018;22(8):675–686. doi:10.1080/14728222.2018.1498845
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
