Back to Journals » Neuropsychiatric Disease and Treatment » Volume 17
The Role of PAX2 in Neurodevelopment and Disease
Authors Lv N, Wang Y, Zhao M, Dong L, Wei H
Received 5 August 2021
Accepted for publication 27 November 2021
Published 7 December 2021 Volume 2021:17 Pages 3559—3567
DOI https://doi.org/10.2147/NDT.S332747
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jun Chen
Na Lv,1 Ying Wang,2 Min Zhao,2 Lina Dong,3 Hongen Wei2
1Department of Physiology, Basic Medical College, Shanxi Medical University, Taiyuan, People’s Republic of China; 2Department of Neurology, Shanxi Provincial People’s Hospital, The Fifth Clinical Medical College of Shanxi Medical University, Taiyuan, People’s Republic of China; 3Central Laboratory, Shanxi Provincial People’s Hospital, The Fifth Clinical Medical College of Shanxi Medical University, Taiyuan, People’s Republic of China
Correspondence: Hongen Wei
Department of Neurology, Shanxi Provincial People’s Hospital, The Fifth Clinical Medical College of Shanxi Medical University, 29 Shuangta Road, Taiyuan, 030012, People’s Republic of China
Tel +86 351 4960173
Fax +86 351 4961994
Email [email protected]
Abstract: In developmental biology, transcription factors are involved in regulating the process of neural development, controlling the differentiation of nerve cells, and affecting the normal functioning of neural circuits. Transcription factors regulate the expression of multiple genes at the same time and have become a key gene category that is recognized to be disrupted in neurodevelopmental disorders such as autism spectrum disorders. This paper briefly introduces the expression and role of PAX2 in neurodevelopment and discusses the neurodevelopmental disorders associated with Pax2 mutations and its possible mechanism. Firstly, mutations in the human Pax2 gene are associated with abnormalities in multiple systems which can result in neurodevelopmental disorders such as intellectual disability, epilepsy and autism spectrum disorders. Secondly, the structure of Pax2 gene and PAX2 protein, as well as the function of Pax2 gene in neural development, was discussed. Finally, a diagram of the PAX2 protein regulatory network was made and a possible molecular mechanism of Pax2 mutations leading to neurodevelopmental disorders from the perspectives of developmental process and protein function was proposed.
Keywords: PAX2, neural development, disorders, mechanism
Introduction
Transcription factors (TFs) play an important role in the developing and adult mammalian brain. In the early stage of embryonic development, TFs determine the expression and relative abundance of guide molecules, mediating progenitor cell differentiation into different types of nerve cells. In the adult stage, TFs are still involved in neurogenesis. The latest research suggests that TFs are responsible for the production of glutamatergic neurons in the adult dentate gyrus of mice.1 Dysregulation of TFs has been implicated in a number of diseases, including neurodevelopmental disorders2–4 and neurodegenerative diseases.5,6 Using viral tools to deliver certain TFs in the mouse brain to reduce the expression of key genes at the transcription level can rescue neuronal damage.4
Here, we focus on paired box 2 (PAX2), a member of the paired box (PAX) transcription factor family expressed mainly in the kidney and central nervous system. The crucial role of the Pax2 genes in brain development and function have become apparent in recent reports on Pax2-related diseases in a Japanese population describing that the patients with Pax2 mutations exhibit developmental abnormalities, including autism spectrum disorders (ASDs), language delay, or mental retardation.7,8 Emerging evidence demonstrates that PAX2 proteins execute important functions during central nervous system development and in adult neurogenesis.9–15 However, the molecular mechanisms required to establish and maintain these functional connections are not yet fully understood. This review examines the role of Pax2 in brain development and the clinical phenotypes related to neurological aspects in patients with Pax2 mutation, focusing on the expression patterns of PAX2 at different stages. A better understanding of these factors will aid in the development of novel treatments to prevent and/or treat diseases associated with Pax2 mutations.
Pax2-Related Neurodevelopmental Disorders
Developmental biology studies have confirmed that the regulation of transcription levels is involved in the process of neurodevelopment, and TFs has become a key gene category that is recognized to be disrupted in neurodevelopmental disorders in the form of a single gene.2 For example, Pax5 and Pax6 in the same gene family as the Pax2 were reported by exome sequencing research to be two candidate risk genes for ASDs,16–18 which is a neurodevelopmental disorder characterized by continuous social interaction and social communication skills deficits and restricted and repetitive behavior patterns, interests or activities.19
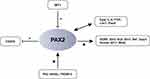
Pax2-related diseases are inherited in an autosomal dominant manner. Patients with Pax2 mutations show renal and eye abnormalities, such as renal dysplasia or optic nerve coloboma,20–22 as well as neurodevelopmental disorders such as ASDs, intellectual disability, epilepsy and developmental delay (Figure 1; Table 1), 7,8,23–30 According to the Leiden Open Variation Database (www.lovd.nl/Pax2), the total number of public variants of the Pax2 gene currently reported is 334, including deletions, insertions, substitutions and duplications of cDNA reference sequences, these variations are mostly located in exons 2, 3, 7, and 8. The most frequently occurring mutation described is c.76dup of exon 2 (previously called c.619insG), which introduces a stop codon 27 amino acids downstream from the homoguanine tract. Nevertheless, the molecular mechanism of how these mutations affect protein function and lead to neurodevelopmental disorders is not clear. Understanding the expression and function of the Pax2 gene may give some clues.
 |
Table 1 List of Pax2 Variants in Neurodevelopmental Disorders |
Nuclear Transcription Factor PAX2
Pax was first isolated and identified through its functions in the Drosophila segmentation gene.31 Based on sequence homology with the Drosophila paired box, nine murine genes containing the paired box have since been isolated, termed Pax1 to Pax9, which together constitute the Pax gene family.32 Throughout the evolutionary process, the Pax gene family has remained highly conserved among different organisms, such as humans, mice, zebrafish33 and chickens.34 The encoded nuclear TFs have a similar structure, with a DNA binding domain of 128 amino acids (paired domain, PD) at the amino-terminal and a transactivation domain (TD) at the carboxyl-terminal. The protein contains a conserved octapeptide (OP) motif and a partial or full homeodomain (HD).35 According to the protein structure, the Pax gene family is divided into four subgroups (Pax1 and Pax9, Pax2, Pax5 and Pax8; Pax3 and Pax7; Pax4 and Pax6).36 The Pax2 gene is a member of the second subgroup, located on mouse chromosome 19 and human chromosome 10q24. In humans, Pax2 consists of 12 exons, with a total length of approximately 70 kb, exons 1–4 encode the PD, exon 5 encodes the OP sequence, and exons 8–11 encode the TD of the PAX2 protein (Figure 1). Mutation or deletion of the OP leads to increased transactivation.37 For example, the deletion of cytosine in exon 5 of patient 13 causes increased gene expression and exhibits seizures (Table 1). The insertion of guanine in exon 2 of patients 3–6, which may cause premature termination of transcription and change the DNA binding function of PD,21 resulting in insufficient PAX2 haplotypes, affecting protein function, and may cause patients to exhibit developmental delays and intellectual disability. While Pax2 consists of 10 exons spanning approximately 91.7 kb in mice, the corresponding translated peptide sequences of human and mouse exon 6 are identical. Studies have shown that the mouse Pax2 gene has 63–67% homology with Drosophila and 69–72% homology with the human gene.38
Alternative splicing is an important mechanism by which eukaryotic genes express a variety of complex proteins. Alternative splicing of PAX2 mRNAs lead to four isoforms in human and mice, including PAX2a, PAX2b, PAX2c, and PAX2d.39–41 PAX2a and PAX2b were first identified in mice, and PAX2c was subsequently discovered in human and mouse kidney. Neither PAX2b nor PAX2c contain exon 6. PAX2c contains an added exon 10 containing 83 bp, causing an exon 11 reading frame migration that leads to the early termination of transcription. PAX2d was also found in the human kidney cDNA library. In this isoform, the first 19 bp of exon 12 are deleted, resulting in the stop codon of the PAX2d transcript being 64 bp downstream of the conventional stop codon. This alternative splicing generates a new reading frame and an extended coding region at the carboxyl terminus, which is widely conserved between human and mice.
Pax2 Gene and Neural Development
Some studies on rodents and human embryos have determined the expression pattern of PAX2 protein in the developmental stage, and the results indicate that expression of the PAX2 protein in the spinal cord and brain during human embryonic development and the description of the role of PAX2 in rodent neurogenesis are very homologous. Pax2 is involved in the early development of mouse embryos, it is expressed in the two borders of the neural plate at E7.5,42 then, neural plate folds to form the neural groove and then closes to generate the neural tube.43 PAX2 is expressed during the development of the neural plate into the neural tube, the neural tube is subdivided into the forebrain, midbrain, hindbrain, and spinal cord.44 Subsequently, PAX2 is expressed in the forebrain,45 midbrain-hindbrain boundary,46 hypothalamus, spinal cord,47 and cerebellum,48 The best studied brain areas in relation to Pax2 are the midbrain-hindbrain, spinal cord, and the cerebellum. The specific function of Pax2 in the hypothalamus is still unclear. The gene dosage of Pax2 and Pax5 in the same subgroup is vital for the function of the organizing center at the midbrain-hindbrain boundary.49 The Pax2 mutant phenotype is strongly influenced by the genetic background of the mouse strain in which it is analyzed. In the C3H/He strain, the hindbrain and cerebellum were completely absent, while in the C57BL/6 strain, brain development was almost normal.50,51 In Pax2 and Pax5 double mutant embryos, the midbrain and hindbrain and cerebellum are absent, however, mutations in Pax2 gene alone do not necessarily affect the production of abnormal phenotypes., suggesting that Pax2 and Pax5 compensate for each other.52
During later embryonic stages (maintenance phase), Pax2, Wnt1 and Fgf8 work together to maintain the midbrain-hindbrain boundary function.49 In the midbrain, the Pax2 expression domain is the source of midbrain dopaminergic neurons.53,54 In the spinal cord and cerebellum, the Pax2 gene was found to be closely related to GABAergic neurons.55 GABAergic interneurons derived from PAX2-positive cells expressed in the cerebellar parenchyma at E13.5 subsequently proliferated to produce different types of inhibitory interneurons, including Golgi and Lugaro cells in the granular layer and basket and stellate cells in the molecular layer.56,57 Studies have shown that PAX2 is expressed in newborn GABAergic neurons on the dorsal spinal cord and is considered a key marker of GABAergic neurons.47 PAX2 acts together with other transcription factors, PTF1A, PRDM13 and LBX1, to participate in the differentiation of GABA precursor neurons and promote the fate of GABAergic neurons,58–61 maintaining continuous expression in GABAergic neurons in adult rats.15 A recent study also concluded that PAX2 is an effective inhibitor of the expression of the Purkinje cell marker FOXP2 and the Purkinje cell differentiation program, and its expression seems to induce the differentiation program characteristics of inhibitory interneurons.62 In our recent research, using CRISPR/Cas9 technology, a Pax2 heterozygous gene deletion mouse (Pax2± KO mice) was constructed. The results of autism-related behavioral tests showed that Pax2± KO mice had significantly increased self-grooming behavior.63 Further study to explore the regulatory mechanism of Pax2 gene deletion leading to autism-like behavior will help to understand the pathogenesis of Pax2-related diseases.
Excitation-inhibitory (E-I) imbalance is considered to be a characteristic of various neurodevelopmental disorders. Large-scale exome sequencing results have revealed that risk genes are enriched in the excitatory and inhibitory neuron lineages.64 GABA is a major inhibitory neurotransmitter widely distributed in the brain and plays an important role in maintaining E-I balance.65–68 It has been reported that GABA or GABA receptor levels in multiple brain regions of children or adults with ASDs are significantly reduced.69–72 Exome sequencing has demonstrated that mutations in the gene that encodes the GABAA receptor subunit disrupt GABA transmission in epilepsy and in people with intellectual disabilities.73–76 Pax2 regulates the cell fate of GABAergic precursor neurons during the development of the cerebellum and spinal cord. Mutation or deletion of Pax2 may lead to impaired transcription regulation, altered GABA levels in the central nervous system, impaired synaptic E-I balance, and neural circuit abnormalities resulting in neurodevelopmental disorders (Figure 2).
Transcriptional Networks Associated with PAX2
The PAX2 recognizes its target gene through the DNA binding function of the pairing domain. Transcription ability is determined by the C-terminal fragment module, which consists of an activation sequence and an inhibitor sequence.77 Therefore, the PAX2 transcription factor acts as both a transcriptional activator and a repressor. The downstream expansion target genes and binding genes of PAX2 have been gradually discovered (Figure 3). It has been reported that GDNF is the target gene of PAX2 in medulloblastoma.78 The complex of transforming factors EYA1 and PAX2 upregulates Six2 and GDNF, which was identified by cDNA microarray analysis. The five genes restricted by PAX2 in the midbrain-hindbrain boundary are En2, Brn1, Sef, Tapp1 and Ncrms.79 PAX2 activates these genes to control the development of the midbrain and cerebellum. Among them, En2 and Brn1 are risk genes for ASDs;80–82 A recent study revealed that PAX2 induces the expression of cyclin D1 by activating AP-1 and promotes the proliferation of colon cancer cells;83 PAX2 activates WNT4 during the development of mammals expression.84 At the same time, PAX2 is also regulated by upstream transcription factors. PAX2 is the direct transcription target of P53;85 the tumor suppressor MENIN inhibits PAX2 through WT1;86 ASH2L participates in the promotion of endometrial cancer progression by upregulating transcription of PAX2.87 During early neurogenesis, PRDM13 regulates and suppresses PAX2, a neuron-specific factor in the neuronal lineage. PAX2 binding sites have also been continuously discovered, including IL-5, PTIP, EYAL, LIM1, PLAC8.87,88 We speculate that Pax2 mutations may affect some factors in the transcriptional network, resulting in abnormal development of the midbrain and hindbrain leading to neurodevelopmental disorders.
Conclusion
In the nervous system, the PAX2 protein participates in the formation of the mid- hindbrain boundary during development, regulates fate determination of precursor neurons, and retains expression in mature cells. However, the specific molecular mechanism of Pax2 regulation remains unclear. Does Pax2 continue to regulate the fate of inhibitory interneurons in the cerebral cortex and other brain regions after development? Which downstream target genes does Pax2 specifically regulate, and how do they function? These questions can be addressed using the latest single-cell transcriptome sequencing technology. Patients with Pax2 mutations exhibit neurodevelopmental disorders, but it is unclear which neurodevelopmental processes affected. The construction of Pax2 conditional gene knockout mice may help to further clarify the pathogenesis of neurodevelopmental disorders in patients with Pax2 mutations.
Author Contributions
Na Lv: Conceptualization, methodology, writing-original draft and review editing. Ying Wang: Writing-review and editing. Min Zhao: Writing-review and editing. Lina Dong: Writing-review and editing. Hongen Wei: Conceptualization, writing-review and editing, funding acquisition, supervision. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81671364), the Natural Science Foundation of Shanxi Province (201801D211010), the Key Medical Science and Technology Project of Shanxi Province (2020XM36), and the Medical Science and Technology Innovation Team of Shanxi Province (2020TD25).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Mateus-Pinheiro A, Alves ND, Patrício P, et al. AP2γ controls adult hippocampal neurogenesis and modulates cognitive, but not anxiety or depressive-like behavior. Mol Psychiatry. 2017;22(12):1725–1734. doi:10.1038/mp.2016.169
2. Teixeira JR, Szeto RA, Carvalho VMA, Muotri AR, Papes F. Transcription factor 4 and its association with psychiatric disorders. Transl Psychiatry. 2021;11(1):19. doi:10.1038/s41398-020-01138-0
3. Estruch SB, Graham SA, Quevedo M, et al. Proteomic analysis of FOXP proteins reveals interactions between cortical transcription factors associated with neurodevelopmental disorders. Hum Mol Genet. 2018;27(7):1212–1227. doi:10.1093/hmg/ddy230
4. Wegmann S, DeVos SL, Zeitler B, et al. Persistent repression of tau in the brain using engineered zinc finger protein transcription factors. Sci Adv. 2021;7(12):eabe1611. doi:10.1126/sciadv.abe1611
5. Hwang JY, Zukin RS. REST, a master transcriptional regulator in neurodegenerative disease. Curr Opin Neurobiol. 2018;48:193–200. doi:10.1016/j.conb.2017.12.008
6. Whitton L, Apostolova G, Rieder D, et al. Genes regulated by SATB2 during neurodevelopment contribute to schizophrenia and educational attainment. PLoS Genet. 2018;14(7):e1007515. doi:10.1371/journal.pgen.1007515
7. Rossanti R, Morisada N, Nozu K, et al. Clinical and genetic variability of PAX2-related disorder in the Japanese population. J Hum Genet. 2020;65(6):541–549. doi:10.1038/s10038-020-0741-y
8. Cheong HI, Cho HY, Kim JH, Yu YS, Ha IS, Choi Y. A clinico-genetic study of renal coloboma syndrome in children. Pediatr Nephrol. 2007;22(9):1283–1289. doi:10.1007/s00467-007-0525-z
9. O’Rahilly R, Müller F. Developmental stages in human embryos: revised and new measurements. Cells Tissues Organs. 2010;192(2):73–84. doi:10.1159/000289817
10. Vinci L, Ravarino A, Fanos V, et al. Immunohistochemical markers of neural progenitor cells in the early embryonic human cerebral cortex. Eur J Histochem. 2016;60(1):2563. doi:10.4081/ejh.2016.2563
11. Wefers AK, Wefers AK, Haberlandt C, et al. Migration of Interneuron Precursors in the Nascent Cerebellar Cortex. The Cerebellum. 2018;17(1):62–71. doi:10.1007/s12311-017-0900-7
12. Scherholz M, Redl E, Wollesen T, de Oliveira AL, Todt C, Wanninger A. Ancestral and novel roles of Pax family genes in mollusks. BMC Evol Biol. 2017;17(1):81. doi:10.1186/s12862-017-0919-x
13. Namm A. Expression of Pax2 protein during the formation of the central nervous system in human embryos. Folia Morphol. 2014;73(3):272–278. doi:10.5603/FM.2014.0043
14. Thompson JA, Ziman M. Pax genes during neural development and their potential role in neuroregeneration. Prog Neurobiol. 2011;95(3):334–351. doi:10.1016/j.pneurobio.2011.08.012
15. Larsson M. Pax2 is persistently expressed by GABAergic neurons throughout the adult rat dorsal horn. Neurosci Lett. 2017;638:96–101. doi:10.1016/j.neulet.2016.12.015
16. O’Roak BJ, Stessman HA, Boyle EA, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi:10.1038/ncomms6595
17. Davis LK, Meyer KJ, Rudd DS, et al. Pax6 3ʹ deletion results in aniridia, autism and mental retardation. Hum Genet. 2008;123(4):371–378. doi:10.1007/s00439-008-0484-x
18. Kikkawa T, Casingal CR, Chun SH, Shinohara H, Hiraoka K, Osumi N. The role of Pax6 in brain development and its impact on pathogenesis of autism spectrum disorder. Brain Res. 2019;1705:95–103. doi:10.1016/j.brainres.2018.02.041
19. Lord C, Brugha TS, Charman T, et al. Autism spectrum disorder. Nat Rev Dis Primers. 2020;6(1):5. doi:10.1038/s41572-019-0138-4
20. Bower MA, Schimmenti LA, Eccles MR, et al. PAX2-Related Disorder. In: Adam MP, Ardinger HH, Pagon RA, editors. GeneReviews(®). University of Washington, Seattle Copyright © 1993–2021; 2018: 154.
21. Galvez-Ruiz A, Lehner AJ, Galindo-Ferreiro A, Schatz P. Three New PAX2 Gene Mutations in Patients with Papillorenal Syndrome. Neuroophthalmology. 2017;41(5):271–278. doi:10.1080/01658107.2017.1307995
22. Schimmenti LA. Renal coloboma syndrome. Eur J Hum Genet. 2011;19(12):1207–1212. doi:10.1038/ejhg.2011.102
23. Cunliffe HE, McNoe LA, Ward TA, Devriendt K, Brunner HG, Eccles MR. The prevalence of PAX2 mutations in patients with isolated colobomas or colobomas associated with urogenital anomalies. J Med Genet. 1998;35(10):806–812. doi:10.1136/jmg.35.10.806
24. Miyazawa T, Nakano M, Takemura Y, et al. A case of renal-coloboma syndrome associated with mental developmental delay exhibiting a novel PAX2 gene mutation. Clin Nephrol. 2009;72(6):497–500. doi:10.5414/cnp72497
25. Deng H, Zhang Y, Xiao H, et al. Diverse phenotypes in children with PAX2‐related disorder. Mol Genet Genomic Med. 2019;7(6):e701. doi:10.1002/mgg3.701
26. Yokoi T, Enomoto Y, Tsurusaki Y, et al. An efficient genetic test flow for multiple congenital anomalies and intellectual disability. Pediatr Int. 2020;62(5):556–561. doi:10.1111/ped.14159
27. Sanyanusin P, Schimmenti LA, McNoe LA, et al. Mutation of the PAX2 gene in a family with optic nerve colobomas, renal anomalies and vesicoureteral reflux. Nat Genet. 1995;9(4):358–364. doi:10.1038/ng0495-358
28. Schimmenti LA, Cunliffe HE, McNoe LA, et al. Further delineation of renal-coloboma syndrome in patients with extreme variability of phenotype and identical PAX2 mutations. Am J Hum Genet. 1997;60(4):869–878.
29. Schimmenti LA, Shim HH, Wirtschafter JD, et al. Homonucleotide expansion and contraction mutations ofPAX2 and inclusion of Chiari 1 malformation as part of Renal-Coloboma syndrome. Hum Mutat. 1999;14(5):369–376. doi:10.1002/(SICI)1098-1004(199911)14:5<369::AID-HUMU2>3.0.CO;2-E
30. Benetti E, Artifoni L, Salviati L, et al. Renal hypoplasia without optic coloboma associated with PAX2 gene deletion. Nephrol Dial Transplant. 2007;22(7):2076–2078. doi:10.1093/ndt/gfm187
31. Deutsch U, Dressler GR, Gruss P. Pax 1, a member of a paired box homologous murine gene family, is expressed in segmented structures during development. Cell. 1988;53(4):617–625. doi:10.1016/0092-8674(88)90577-6
32. Stapleton P, Weith A, Urbánek P, Kozmik Z, Busslinger M. Chromosomal localization of seven PAX genes and cloning of a novel family member, PAX-9. Nat Genet. 1993;3(4):292–298. doi:10.1038/ng0493-292
33. Krauss S, Johansen T, Korzh V, Fjose A. Expression pattern of zebrafish pax genes suggests a role in early brain regionalization. Nature. 1991;353(6341):267–270. doi:10.1038/353267a0
34. Goulding MD, Lumsden A, Gruss P. Signals from the notochord and floor plate regulate the region-specific expression of two Pax genes in the developing spinal cord. Development. 1993;117(3):1001–1016. doi:10.1242/dev.117.3.1001
35. Walther C, Guenet JL, Simon D, et al. Pax: a murine multigene family of paired box-containing genes. Genomics. 1991;11(2):424–434. doi:10.1016/0888-7543(91)90151-4
36. Dahl E, Koseki H, Balling R. Pax genes and organogenesis. Bioessays. 1997;19(9):755–765. doi:10.1002/bies.950190905
37. Lechner MS, Dressler GR. Mapping of Pax-2 transcription activation domains. J Biol Chem. 1996;271(35):21088–21093. doi:10.1074/jbc.271.35.21088
38. Dressler GR, Deutsch U, Chowdhury K, Nornes HO, Gruss P. Pax2, a new murine paired-box-containing gene and its expression in the developing excretory system. Development. 1990;109(4):787–795. doi:10.1242/dev.109.4.787
39. Ritz-Laser B, Estreicher A, Gauthier B, Philippe J. The paired homeodomain transcription factor Pax-2 is expressed in the endocrine pancreas and transactivates the glucagon gene promoter. J Biol Chem. 2000;275(42):32708–32715. doi:10.1074/jbc.M005704200
40. Tavassoli K, Rüger W, Horst J. Alternative splicing in PAX2 generates a new reading frame and an extended conserved coding region at the carboxy terminus. Hum Genet. 1997;101(3):371–375. doi:10.1007/s004390050644
41. Ward TA, Nebel A, Reeve AE, Eccles MR. Alternative messenger RNA forms and open reading frames within an additional conserved region of the human PAX-2 gene. Cell Growth Differ. 1994;5(9):1015–1021.
42. Rowitch DH, McMahon AP. Pax-2 expression in the murine neural plate precedes and encompasses the expression domains of Wnt-1 and En-1. Mech Dev. 1995;52(1):3–8. doi:10.1016/0925-4773(95)00380-j
43. Frisen J, Johansson CB, Lothian C, Lendahl U. Central nervous system stem cells in the embryo and adult. Cell Mol Life Sci. 1998;54(9):935–945. doi:10.1007/s000180050224
44. Schwarz M, Alvarez-Bolado G, Dressler G, Urbanek P, Busslinger M, Gruss P. Pax2/5 and Pax6 subdivide the early neural tube into three domains. Mech Dev. 1999;82(1–2):29–39. doi:10.1016/s0925-4773(99)00005-2
45. Fotaki V, Price DJ, Mason JO. Newly identified patterns of Pax2 expression in the developing mouse forebrain. BMC Dev Biol. 2008;8(1):79. doi:10.1186/1471-213X-8-79
46. Pfeffer PL, Bouchard M, Busslinger M. Pax2 and homeodomain proteins cooperatively regulate a 435 bp enhancer of the mouse Pax5 gene at the midbrain-hindbrain boundary. Development. 2000;127(5):1017–1028. doi:10.1242/dev.127.5.1017
47. Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Lhx1 and Lhx5 maintain the inhibitory-neurotransmitter status of interneurons in the dorsal spinal cord. Development. 2007;134(2):357–366. doi:10.1242/dev.02717
48. Maricich SM, Herrup K. Pax-2 expression defines a subset of GABAergic interneurons and their precursors in the developing murine cerebellum. J Neurobiol. 1999;41(2):281–294. doi:10.1002/(sici)1097-4695(19991105)41:2<281::aid-neu10>3.0.co;2-5
49. Rhinn M, Brand M. The midbrain–hindbrain boundary organizer. Curr Opin Neurobiol. 2001;11(1):34–42. doi:10.1016/s0959-4388(00)00171-9
50. Favor J, Sandulache R, Neuhäuser-Klaus A, et al. The mouse Pax2(1Neu) mutation is identical to a human PAX2 mutation in a family with renal-coloboma syndrome and results in developmental defects of the brain, ear, eye, and kidney. Proc Natl Acad Sci U S A. 1996;93(24):13870–13875. doi:10.1073/pnas.93.24.13870
51. Schwarz M, Alvarez-Bolado G, Urbánek P, Busslinger M, Gruss P. Conserved biological function between Pax-2 and Pax-5 in midbrain and cerebellum development: evidence from targeted mutations. Proc Natl Acad Sci U S A. 1997;94(26):14518–14523. doi:10.1073/pnas.94.26.14518
52. Bouchard M, Pfeffer P, Busslinger M. Functional equivalence of the transcription factors Pax2 and Pax5 in mouse development. Development. 2000;127(17):3703–3713. doi:10.1242/dev.127.17.3703
53. Simon HH, Bhatt L, Gherbassi D, Sgadó P, Alberí L. Midbrain dopaminergic neurons: determination of their developmental fate by transcription factors. Ann N Y Acad Sci. 2003;991:36–47. doi:10.1111/j.1749-6632.2003.tb07461.x
54. Niu Y, Moghimyfiroozabad S, Moghimyfiroozabad A. The factors for the early and late development of midbrain dopaminergic neurons segregate into two distinct evolutionary clusters. Brain Disorders. 2021;100002. doi:10.1016/j.dscb.2021.100002
55. Borromeo MD, Meredith DM, Castro DS, et al. A transcription factor network specifying inhibitory versus excitatory neurons in the dorsal spinal cord. Development. 2014;141(14):2803–2812. doi:10.1242/dev.105866
56. Leto K, Carletti B, Williams IM, Magrassi L, Rossi F. Different types of cerebellar GABAergic interneurons originate from a common pool of multipotent progenitor cells. J Neurosci. 2006;26(45):11682–11694. doi:10.1523/jneurosci.3656-06.2006
57. Fauquier T, Romero E, Picou F, et al. Severe impairment of cerebellum development in mice expressing a dominant-negative mutation inactivating thyroid hormone receptor alpha1 isoform. Dev Biol. 2011;356(2):350–358. doi:10.1016/j.ydbio.2011.05.657
58. Huang M, Huang T, Xiang Y, et al. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol. 2008;322(2):394–405. doi:10.1016/j.ydbio.2008.06.031
59. Chang JC, Meredith DM, Mayer PR, et al. Prdm13 mediates the balance of inhibitory and excitatory neurons in somatosensory circuits. Dev Cell. 2013;25(2):182–195. doi:10.1016/j.devcel.2013.02.015
60. Guo Z, Zhao C, Huang M, et al. Tlx1/3 and Ptf1a control the expression of distinct sets of transmitter and peptide receptor genes in the developing dorsal spinal cord. J Neurosci. 2012;32(25):8509–8520. doi:10.1523/jneurosci.6301-11.2012
61. Mona B, Uruena A, Kollipara RK, et al. Repression by PRDM13 is critical for generating precision in neuronal identity. Elife. 2017;6:e25787. doi:10.7554/eLife.25787
62. Lowenstein ED, Rusanova A, Stelzer J, et al. Olig3 regulates early cerebellar development. Elife. 2021;10:e64684. doi:10.7554/eLife.64684
63. Wei H, Wang M, Lv N, et al. Increased repetitive self-grooming occurs in Pax2 mutant mice generated using CRISPR/Cas9. Behav Brain Res. 2020;393:112803. doi:10.1016/j.bbr.2020.112803
64. Satterstrom FK, Kosmicki JA, Wang J, et al. Large-Scale Exome Sequencing Study Implicates Both Developmental and Functional Changes in the Neurobiology of Autism. Cell. 2020;180(3):568–584.e23. doi:10.1016/j.cell.2019.12.036
65. Sohal VS, Rubenstein JLR. Excitation-inhibition balance as a framework for investigating mechanisms in neuropsychiatric disorders. Mol Psychiatry. 2019;24(9):1248–1257. doi:10.1038/s41380-019-0426-0
66. Di J, Li J, O’Hara B, et al. The role of GABAergic neural circuits in the pathogenesis of autism spectrum disorder. Int J Dev Neurosci. 2020;80(2):73–85. doi:10.1002/jdn.10005
67. Yizhar O, Fenno LE, Prigge M, et al. Neocortical excitation/inhibition balance in information processing and social dysfunction. Nature. 2011;477(7363):171–178. doi:10.1038/nature10360
68. Bak LK, Schousboe A, Waagepetersen HS. The glutamate/GABA-glutamine cycle: aspects of transport, neurotransmitter homeostasis and ammonia transfer. J Neurochem. 2006;98(3):641–653. doi:10.1111/j.1471-4159.2006.03913.x
69. Fatemi SH, Folsom TD, Reutiman TJ, Thuras PD. Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum. 2009;8(1):64–69. doi:10.1007/s12311-008-0075-3
70. Oblak AL, Gibbs TT, Blatt GJ. Decreased GABA(B) receptors in the cingulate cortex and fusiform gyrus in autism. J Neurochem. 2010;114(5):1414–1423. doi:10.1111/j.1471-4159.2010.06858.x
71. Fung LK, Flores RE, Gu M, et al. Thalamic and prefrontal GABA concentrations but not GABA(A) receptor densities are altered in high-functioning adults with autism spectrum disorder. Mol Psychiatry. 2021;26(5):1634–1646. doi:10.1038/s41380-020-0756-y
72. Puts NAJ, Wodka EL, Harris AD, et al. Reduced GABA and altered somatosensory function in children with autism spectrum disorder. Autism Res. 2017;10(4):608–619. doi:10.1002/aur.1691
73. Zapata J, Moretto E, Hannan S, et al. Epilepsy and intellectual disability linked protein Shrm4 interaction with GABA(B)Rs shapes inhibitory neurotransmission. Nat Commun. 2017;8:14536. doi:10.1038/ncomms14536
74. Chuang SH, Reddy DS. Genetic and Molecular Regulation of Extrasynaptic GABA-A Receptors in the Brain: therapeutic Insights for Epilepsy. J Pharmacol Exp Ther. 2018;364(2):180–197. doi:10.1124/jpet.117.244673
75. Butler KM, Moody OA, Schuler E, et al. De novo variants in GABRA2 and GABRA5 alter receptor function and contribute to early-onset epilepsy. Brain. 2018;141(8):2392–2405. doi:10.1093/brain/awy171
76. May P, Girard S, Harrer M, et al. Rare coding variants in genes encoding GABA(A) receptors in genetic generalised epilepsies: an exome-based case-control study. Lancet Neurol. 2018;17(8):699–708. doi:10.1016/s1474-4422(18)30215-1
77. Dörfler P, Busslinger M. C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8. EMBO j. 1996;15(8):1971–1982. doi:10.1002/j.1460-2075.1996.tb00548.x
78. Reidy KJ, Rosenblum ND. Cell and molecular biology of kidney development. Semin Nephrol. 2009;29(4):321–337. doi:10.1016/j.semnephrol.2009.03.009
79. Bouchard M, Grote D, Craven SE, Sun Q, Steinlein P, Busslinger M. Identification of Pax2-regulated genes by expression profiling of the mid-hindbrain organizer region. Development. 2005;132(11):2633–2643. doi:10.1242/dev.01833
80. Hnoonual A, Sripo T, Limprasert P. Whole-exome sequencing identifies a novel heterozygous missense variant of the EN2 gene in two unrelated patients with autism spectrum disorder. Psychiatr Genet. 2016;26(6):297–301. doi:10.1097/ypg.0000000000000153
81. Boschian C, Messina A, Bozza A, et al. Impaired Neuronal Differentiation of Neural Stem Cells Lacking the Engrailed-2 Gene. Neuroscience. 2018;386:137–149. doi:10.1016/j.neuroscience.2018.06.032
82. Snijders Blok L, Kleefstra T, Venselaar H, et al. De Novo Variants Disturbing the Transactivation Capacity of POU3F3 Cause a Characteristic Neurodevelopmental Disorder. Am J Hum Genet. 2019;105(2):403–412. doi:10.1016/j.ajhg.2019.06.007
83. Zhang HS, Yan B, Li XB, et al. PAX2 protein induces expression of cyclin D1 through activating AP-1 protein and promotes proliferation of colon cancer cells. J Biol Chem. 2012;287(53):44164–44172. doi:10.1074/jbc.M112.401521
84. Torban E, Dziarmaga A, Iglesias D, et al. PAX2 activates WNT4 expression during mammalian kidney development. J Biol Chem. 2006;281(18):12705–12712. doi:10.1074/jbc.M513181200
85. Saifudeen Z, Liu J, Dipp S, et al. A p53-Pax2 pathway in kidney development: implications for nephrogenesis. PLoS One. 2012;7(9):e44869. doi:10.1371/journal.pone.0044869
86. Xu B, Zeng DQ, Wu Y, et al. Tumor suppressor menin represses paired box gene 2 expression via Wilms tumor suppressor protein-polycomb group complex. J Biol Chem. 2011;286(16):13937–13944. doi:10.1074/jbc.M110.197830
87. Zeng K, Wu Y, Wang C, et al. ASH2L is involved in promotion of endometrial cancer progression via upregulation of PAX2 transcription. Cancer Sci. 2020;111(6):2062–2077. doi:10.1111/cas.14413
88. Liu P, Gao Y, Huan J, et al. Upregulation of PAX2 promotes the metastasis of esophageal cancer through interleukin-5. Cell Physiol Biochem. 2015;35(2):740–754. doi:10.1159/000369734
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.