Back to Journals » OncoTargets and Therapy » Volume 12
The role of miR-382-5p in glioma cell proliferation, migration and invasion
Authors Wang J, Chen C, Yan X , Wang P
Received 28 November 2018
Accepted for publication 2 May 2019
Published 25 June 2019 Volume 2019:12 Pages 4993—5002
DOI https://doi.org/10.2147/OTT.S196322
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Jinjin Wang, Chunfeng Chen, Xu Yan, Peng Wang
Department of Neurosurgery, The Affiliated Jiangsu Shengze Hospital of Nanjing Medical University, Suzhou, Jiangsu, 215000, People’s Republic of China
Background: Dysregulation of a single miRNA can play an essential role in tumor development and progression. Recent studies have shown that miR-382-5p can function as an oncogene or as a tumor suppressor in different types of cancers. However, the role of miR-382-5p in glioma growth and expansion has not been characterized.
Methods: Quantitative real time-PCR (qRT-PCR) was used to measure miR-382-5p levels in glioma tissues. The miR-382-5p mimics and inhibitors were employed to upregulate and downregulate miR-382-5p expression respectively in glioma cells. EdU assay was used to assess cell proliferation. Wound healing and Transwell assays were employed to evaluate cell migration and invasion. Western blot was used to measure the changes of epithelial-to-mesenchymal transition (EMT) markers and the potential miR-382-5p target genes.
Results: We found that miR-382-5p levels were low in glioma tissues as determined by qRT-PCR. EdU assay showed that upregulation of miR-382-5p significantly decreased cell proliferation in both U87 and U251 cells. Wound healing rate was significantly decreased in response to miR-382-5p mimics and significantly increased in response to miR-382-5p inhibitors. Transwell migration assays further confirmed the inhibitory effects of miR-382-5p on the migration in both U251 and U87 cells. Transwell invasion assays showed that upregulation of miR-382-5p resulted in a remarkable decrease in the number of invading cells, whereas downregulation of miR-382-5p led to a significant increase in the numbers of invading U87 and U251 cells. In addition, overexpression of miR-382-5p decreased the protein levels of N-cadherin, Snail and Slug, and increased E-cadherin levels, in glioma cells. Furthermore, miR-382-5p levels negatively correlated with Y box-binding protein 1 (YBX1) in lower grade glioma tissues, and negatively regulated the expression of YBX1 in glioma cells.
Conclusion: In summary, miR-382-5p inhibited proliferation, migration, invasion, and the EMT in glioma cells, possibly through targeting the oncogene YBX1.
Keywords: miR-382, glioma, proliferation, migration, invasion
Introduction
Glioma is the most common malignant tumor in the central nervous system. It has a poor prognosis and a high recurrence rate. Despite significant advances in surgery, chemotherapy, and radiotherapy, the mean survival time of patients with malignant gliomas has not significantly improved.1 The 5-year survival rate is below 10%, and the average time from diagnosis to death is less than 18 months.2 Therefore, identification of novel biomarkers and therapeutic targets for glioma is critical.
MicroRNAs (miRNAs) are a family of small non-coding RNAs with approximately 18–22 nucleotides in the mature form. They post-transcriptionally control other RNAs by binding to the 3ʹ untranslated regions (UTRs) of their targets.3 MiRNAs are thought to regulate more than 70% of human genes, and a single miRNA can target hundreds of genes.4,5 Dysregulation of a single miRNA can play an essential role in tumor development and progression.4,5 MiRNAs can function as either oncogenes or tumor suppressor genes, depending on the target genes that they regulate.6,7
MiR-382, a microRNAs in the chromosome 14q32 locus, has been shown to be involved in development, metastasis, and therapeutic resistance in multiple types of cancers. It functions as a tumor suppressor in colorectal cancer,8,9 ovarian cancer,10 esophageal squamous cell carcinoma,11,12 osteosarcoma,13,14 prostate cancer,15 melanoma,16 and non-small cell lung cancer.17 MiR-382 exhibits oncogenic properties in gastric cancer,18 hepatocellular carcinoma,19 and breast cancer.20 These differences indicate that miR-382 may exert its role in cell- and tissue-specific manners.
MiRNA dysregulation in glioma has been extensively studied.21 Levels of miRNAs in cerebrospinal fluid and brain tissue in patients with glioma are reliable markers, and may have diagnostic value for glioma.22 The role of miR-382-5p in glioma cell growth and progression has not been characterized. In the present study, we transfected miR-382-5p mimics or inhibitors into U251 and U87 glioma cells to determine the effects of this miRNA on glioma cell proliferation, migration, invasion, and epithelial to mesenchymal transition (EMT). In addition, we found that Y box-binding protein 1 (YBX1) expression was negatively regulated by miR-382-5p, which may be responsible for the role of miR-382-5p in glioma cells.
Materials and methods
Patients and samples
All glioma and nontumor tissue specimens were collected from the First Affiliated Hospital of Soochow University, Suzhou, China. All patients were naïve to immunotherapy, radiation, and chemotherapy. Fresh samples were stored at −80 °C immediately after surgical removal. Written informed consent was obtained from each subject or legal guardian to use the specimens for future investigations and research purposes. All data were analyzed anonymously and all experiments were performed in compliance with the Helsinki Declaration. This study was approved by the Ethics Committee of Soochow University.
Cell lines and cell culture
The HEK293T cell line and human glioma cell lines were purchased from the Shanghai Cell Bank, Type Culture Collection Committee, Chinese Academy of Sciences. The cells were grown in DMEM supplemented with 10% fetal bovine serum (Gibco). All cell lines were cultured in a cell incubator with a 5% CO2 atmosphere under saturated humidity at 37 °C.
MiRNA transfection
MiRNA-382-5p mimics, inhibitors, and the corresponding negative controls (NC) were synthesized by Biomics Biotech (Nantong, China). The cells were grown to 70–90% confluence in six-well plates, then transfected with the purchased oligonucleotides using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
RNA extraction and quantitative real time-PCR (qRT-PCR)
Total RNA was extracted from tissues or cells with TRIzol (Invitrogen, Waltham, MA, USA) according to the manufacturer’s protocol. To detect miR-382-5p, reverse transcription was performed using a cDNA Reverse transcription kit (Roche) and a miR-382-5p-specific reverse transcription primer (Biomics Biotech), and qPCR was performed using a SYBR Green PCR Master kit (Roche) and a miR‑382-5p qPCR Detection Primer Set (Biomics Biotech). To detect U6 snRNA, an internal control, the extracted RNA was reversely transcribed into cDNA with the cDNA Reverse transcription kit (Roche), and U6 snRNA was amplified with a SYBR Green PCR Master kit (Roche) and a mixture of forward and reverse primers for U6 (Forward: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ; Reverse: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ). qPCR temperature cycles were performed as follows: 50 °C for 2 min and 95 °C for 10 min; 45 cycles at 95 °C for 15 s and 60 °C for 1 min. Data were acquired and processed automatically by the Applied Biosystems 7500 Real-Time PCR System. The expression of miR-382-5p relative to U6 snRNA was calculated according to a previous method for statistics.23
EdU assay
The 5-ethynyl-20-deoxyuridine (EdU) incorporation assay was performed using an EdU assay kit (Ribobio, Guangzhou, China) according to the manufacturer’s instructions. Briefly, cells were exposed to 50 μM EdU for 2 h, then fixed with 4% formaldehyde. The cells were then treated with 2 mg/mL glycine to neutralize the formaldehyde, then permeabilized with 0.5% Triton X-100. Finally, the cells were reacted with 100 μL of 1X Apollo reaction cocktail for 30 min, followed by incubation with 100 μL of Hoechst 33,342 (5 μg/mL). Images were acquired using an Olympus IX-71 inverted microscope (Tokyo, Japan). The percentage of EdU-positive cells was calculated by dividing the number of EdU-positive cells by the number of Hoechst-stained cells.
Wound healing assay
Cell migration behavior was evaluated using the wound healing assay as previously described.24 Briefly, monolayers were wounded with a plastic pipette tip, the dead cells were removed by rinsing, and serum-free media was added. At the designated times (0 h, 12 h, and 24 h), images of three randomly selected fields at the lesion border were acquired using an Olympus IX-71 inverted microscope. The wound healing rate was calculated based on the captured images.
Transwell migration and invasion assays
Cell migration and invasion assays were performed using a transwell system with a polycarbonate filter membrane as previously described.25 To assess invasion ability, the filters were precoated with 10 μg of Matrigel (BD, Franklin Lakes, NJ, USA). Five fields of adherent cells in each well were randomly photographed using an Olympus IX71 inverted microscope. The number of migrating or invading cells was counted in the captured images.
Protein extraction and Western blot
Total protein was extracted from the cultured cells, and protein concentrations were determined using a BCA Protein Assay Kit (Beyotime, Haimen, China). Equal amounts of total protein were used for Western blot analysis similar to a previously described protocol.24 The following primary antibodies were purchased from Abcam (Cambridge, UK): N-cadherin (1:2000), E-cadherin (1:1000), and Snail and Slug (1:2000). The KLF12 (1:800) was purchased from Proteintech (Wuhan, China). The YBX1 (1:500) and β-Actin (1:1500) antibodies were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Band densities were analyzed using Image J software (National Institute of Health, Bethesda, MD, USA). Relative protein levels were determined by normalizing the densitometry value of the proteins of interest to that of β-Actin.
Statistical analysis
The miR-382-5p levels in glioma tissues were presented as scatter plots with the median as the measure of center. Differences between the nontumor group and the glioma subgroups were evaluated using the Kruskal–Wallis test and the Mann–Whitney U test. In vitro experiments were repeated at least three times, and the data are expressed as the means ± S.E.M. Comparisons between two groups were performed using Student’s t-test, and the differences among three groups were determined using one-way analysis of variance (ANOVA) followed by Dunnett’s post hoc test. Statistical analyses were performed using SPSS version 13.0 (SPSS Inc., Chicago, IL, USA). Tests were 2-tailed and P<0.05 was considered statistically significant.
Results
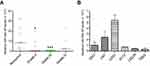
Changes in miR-382-5p levels in glioma tissues and cell lines
We used miR-382-5p specific primers for qRT-PCR analysis in nontumor and glioma tissues. As shown in Figure 1A, miR-382-5p levels were lower in Grade II (n=9, P=0.039), Grade III (n=8, P=0.002), and Grade IV (n=7, P=0.142) glioma tissues compared to those in nontumor brain tissues (n=7). In addition, qRT-PCR was performed in HEK293T cells and different glioma cell lines, and we found that miR-382-5p levels was relative high in U87 and U251 cells (Figure 1B).
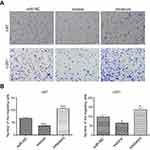
Effects of up- or downregulation of miR-382-5p on glioma cell proliferation
Transfection with miR-382-5p mimics resulted in a significant decrease in the percentage of EdU-positive cells in both U87 (P=0.001) and U251 (P=0.034) cells compared to transfection with miR-NC. In contrast, transfection with miR-382-5p inhibitors did not significantly affect the percentage of EdU-positive U87 or U251 cells (Figure 2). These results suggested that upregulation of miR-382-5p played an inhibitory role in glioma cell proliferation.
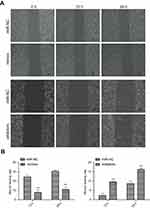
Effects of up- or downregulation of miR-382-5p on glioma cell migration
The wound healing rate was significantly decreased in the miR-382-5p mimics group (P=0.013 at both 12 h and 24 h), and was significantly increased in the miR-382-5p inhibitors group (P=0.006 at 12 h and P=0.025 at 24 h) following wound injury (Figure 3). Moreover, transwell migration assay showed that transfection with miR-382-5p mimics significantly decreased the number of U87 (P<0.001) and U251 (P=0.036) cells that migrated to the chamber. Transfection with miR-382-5p inhibitors resulted in significant increases in U87 and U251 cells (P<0.001 for U87 and P=0.023 for U251) (Figure 4). These findings indicated that miR-382-5p played an inhibitory role in glioma cell migration.
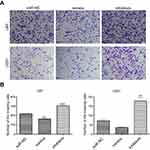
Effects of up- or downregulation of miR-382-5p on glioma cell invasion
We used the transwell assay in the presence of Matrigel to determine the effects of up- or downregulation of miR-382-5p on glioma cell invasion. In U87 cells (Figure 5), upregulation of miR-382-5p by its mimics resulted in a significant decrease in the number of cells that passed through the Matrigel (P=0.001), whereas downregulation of miR-382-5p by its inhibitors led to a significant increase in the number of invading cells (P<0.001). In U251 cells (Figure 5), the number of invading cells showed a trend toward a decrease in the miR-382-5p mimics group (P=0.146), and the number of invading cells was significantly increased in the miR-382-5p inhibitors group (P=0.002). These data suggested that miR-382-5p played an inhibitory role in glioma cell invasion.
Effects of up- or downregulation of miR-382-5p on the EMT in glioma cells
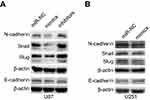
The epithelial-to-mesenchymal transition (EMT) has been identified as a key regulator of the invasion of glioma cells.26 Western blot was used to detect important signaling molecules that mediate the EMT process in glioma cells. Transfection with miR-382-5p mimics decreased mesenchymal markers such as N-cadherin, and Snail and Slug, whereas transfection with miR-382-5p inhibitors increased the protein levels of these mesenchymal markers in U87 cells (Figure 6A). No differences in E-cadherin were observed between mimics and miR-NC or between inhibitors and miR-NC in U87 cells (Figure 6A). Transfection with miR-382-5p mimics resulted in increased E-cadherin levels, and decreased N-cadherin, and Snail and Slug levels, in U251 cells (Figure 6B). These results indicated that miR-382-5p may regulate the EMT process in glioma cells.
Analysis of the relationship of miR-382-5p with its target genes in glioma
Previous studies have identified several target genes of miR-382-5p. We performed co-expression analyses between these genes and miR-382-5p in tissues of patients with LGG from the TCGA consortium (Table 1). Patient data and gene expression datasets were obtained from StarBase (
 | Table 1 Correlation analyses of miR-382-5p with its target genes in lower grade glioma tissues |
Discussion
In this study, we demonstrated that overexpression of miR-382-5p inhibited glioma cell proliferation, migration, and invasion of glioma cells, whereas inhibition of miR-382-5p resulted in opposite effects. These findings were consistent with those in previous studies of colorectal cancer,8,9 ovarian cancer,10 esophageal squamous cell carcinoma (ESCC),11,12 osteosarcoma,13,14 prostate cancer,15 melanoma,16 and non-small cell lung cancer (NSCLC).17 However, other reports have shown an oncogenic function of miR-382-5p in cancer cell progression. For example, inhibition of miR-382 expression reduced xenograft tumor growth and microvessel density in gastric cancer,18 and promoted breast cancer cell proliferation, survival, migration, invasion, and in vivo tumorigenesis/metastasis.20 MiR-382-5p inhibitors neutralized the effects of hepatitis B virus core protein on the migration and invasion ability of liver cancer cells.19 These studies indicated that the role of miR-382-5p in tumor growth and metastasis is heterogeneous and is cell- and tissue-dependent.
The EMT is the process by which epithelial cells transform into mesenchymal cells, which is a vital step in invasion of glioma.26 The EMT process is characterized by loss of epithelial markers and gain of mesenchymal markers. Western blot analysis of the expression of the epithelial marker E-cadherin and the mesenchymal markers N-cadherin, and Snail and Slug in glioma cells showed that miR-382-5p inhibited the EMT process. Thus, our findings and those from previous studies of cancer cells in osteosarcoma, ovarian cancer, and esophageal squamous cell carcinoma consistently showed the suppressive effects of miR-382-5p on the EMT,10,11,14 which might be responsible for its inhibitory effects on tumor cell migration and invasion.
MiRNAs play different roles by targeting different genes. Therefore, investigation of the target genes of miRNAs is critical to understanding the molecular mechanisms by which a miRNA promotes or suppresses oncogenesis. MiR-382-5p has been shown to directly target several genes including Y box-binding protein 1 (YBX1),14 receptor tyrosine kinase like orphan receptor 1 (ROR1),10 krueppel-like factor 12 (KLF12),8 homeodomain-interacting protein kinase 3 (HIPK3),8 nuclear receptor subfamily 2 group F member 2 (NR2F2, also known as COUP-TFII),9,15 and histone-lysine N-methyltransferase (KMT5A, also known as SETD8).17,28 Analysis of co-expression of these genes and miR-382-5p showed that KLF12 and YBX1 significantly negatively correlated with miR382-5p in patients with LGG. Moreover, we found that transfection with miR-382-5p mimics resulted in downregulation of the expression of YBX1 in glioma cells, but did not significantly affect KLF12 protein levels. A previous study showed that overexpression of YBX1 promoted drug resistance to temozolomide and increased the growth viability of glioma cells.29 Therefore, miR-382-5p may target YBX1, resulting in glioma growth suppression.
We found that miR-382-5p levels were decreased in various grades of glioma tissues, which was consistent with downregulation of miR-382-5p in other types of cancers including osteosarcoma,13 ovarian cancer,10 colorectal cancer,8,9 prostate cancer,15 and NSCLC.17 Downregulation of miR-382-5p was an indicator of poor outcome, relapse, or metastasis in osteosarcoma and ESCC.12,14 In contrast, higher miR-382-5p levels were significantly associated with worse overall survival and disease-free survival of patients with breast cancer, which was consistent with the oncogenic function of miR-382-5p in breast cancer cells.20 These connections indicated the potential diagnostic and prognostic value of miR-382-5p for these cancers. Further studies are needed to characterize the relationship between miR-382-5p levels and survival of patients with glioma.
Our study suffered from the following limitations. We demonstrated an inhibitory role for miR-382-5p in glioma cell progression in immortalized cell lines such U251 and U87. These studies should be repeated using patient-derived cells in the future. In addition, glioma orthotopic xenograft experiments would help to confirm the suppressive effects of miR-382-5p on glioma cell growth in vivo.
Conclusions
In the present study, we found that miR-382-5p levels were low in glioma tissues, and that upregulation of miR-382-5p inhibited cell proliferation, migration, invasion, and the EMT process in glioma cells. MiR-382-5p may function as a tumor suppressor in glioma progression by negatively regulating the oncogene YBX1.
Acknowledgment
This study was supported by the Program of Science and Education to Promote Health of Wujiang District, Suzhou, People’s Republic of China (grant number WWK201714).
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64(6):479–489. doi:10.1093/jnen/64.6.479
2. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi:10.1016/S1470-2045(09)70025-7
3. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
4. Bhattacharya A, Ziebarth JD, Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41(Databaseissue):D977–D982. doi:10.1093/nar/gks1138
5. Miranda KC, Huynh T, Tay Y, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126(6):1203–1217. doi:10.1016/j.cell.2006.07.031
6. Huang J, Zhang SY, Gao YM, et al. MicroRNAs as oncogenes or tumour suppressors in oesophageal cancer: potential biomarkers and therapeutic targets. Cell Prolif. 2014;47(4):277–286. doi:10.1111/cpr.12109
7. Mamoori A, Gopalan V, Lam AK. Role of miR-193a in cancer: complexity and factors control the pattern of its expression. Curr Cancer Drug Targets. 2018;18(7):618–628. doi:10.2174/1568009618666180308105727
8. Yao H, Xia D, Li ZL, et al. MiR-382 functions as tumor suppressor and chemosensitizer in colorectal cancer. Biosci Rep. 2018. doi:10.1042/BSR20180441
9. Zhou B, Song J, Han T, et al. MiR-382 inhibits cell growth and invasion by targeting NR2F2 in colorectal cancer. Mol Carcinog. 2016;55(12):2260–2267. doi:10.1002/mc.22466
10. Tan H, He Q, Gong G, et al. miR-382 inhibits migration and invasion by targeting ROR1 through regulating EMT in ovarian cancer. Int J Oncol. 2016;48(1):181–190. doi:10.3892/ijo.2015.3241
11. Feng J, Qi B, Guo L, et al. miR-382 functions as a tumor suppressor against esophageal squamous cell carcinoma. World J Gastroenterol. 2017;23(23):4243–4251. doi:10.3748/wjg.v23.i23.4243
12. Qi B, Lu JG, Yao WJ, et al. Downregulation of microRNA-382 is associated with poor outcome of esophageal squamous cell carcinoma. World J Gastroenterol. 2015;21(22):6884–6891. doi:10.3748/wjg.v21.i22.6884
13. Xu M, Jin H, Xu CX, et al. miR-382 inhibits tumor growth and enhance chemosensitivity in osteosarcoma. Oncotarget. 2014;5(19):9472–9483. doi:10.18632/oncotarget.2418
14. Xu M, Jin H, Xu CX, et al. miR-382 inhibits osteosarcoma metastasis and relapse by targeting Y box-binding protein 1. Mol Ther. 2015;23(1):89–98. doi:10.1038/mt.2014.197
15. Zhang W, Liu J, Qiu J, et al. MicroRNA-382 inhibits prostate cancer cell proliferation and metastasis through targeting COUP-TFII. Oncol Rep. 2016;36(6):3707–3715. doi:10.3892/or.2016.5141
16. Hanniford D, Segura MF, Zhong J, et al. Identification of metastasis-suppressive microRNAs in primary melanoma. J Natl Cancer Inst. 2015;107(3). doi:10.1093/jnci/dju494
17. Chen T, Ren H, Thakur A, et al. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 2017;86:248–253. doi:10.1016/j.biopha.2016.12.007
18. Seok JK, Lee SH, Kim MJ, Lee YM. MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic Acids Res. 2014;42(12):8062–8072. doi:10.1093/nar/gku515
19. Du J, Bai F, Zhao P, et al. Hepatitis B core protein promotes liver cancer metastasis through miR-382-5p/DLC-1 axis. Biochim Biophys Acta Mol Cell Res. 2018;1865(1):1–11. doi:10.1016/j.bbamcr.2017.09.020
20. Ho JY, Hsu RJ, Liu JM, et al. MicroRNA-382-5p aggravates breast cancer progression by regulating the RERG/Ras/ERK signaling axis. Oncotarget. 2017;8(14):22443–22459. doi:10.18632/oncotarget.12338
21. Wang S, Yin Y, Liu S. Roles of microRNAs during glioma tumorigenesis and progression. Histol Histopathol. 2018;34(3):213–222. doi:10.14670/HH-18-040
22. Zhou Q, Liu J, Quan J, Liu W, Tan H, Li W. MicroRNAs as potential biomarkers for the diagnosis of glioma: A systematic review and meta-analysis. Cancer Sci. 2018;109(9):2651–2659. doi:10.1111/cas.13714
23. Chen C, Ridzon DA, Broomer AJ, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33(20):e179. doi:10.1093/nar/gni178
24. Zhang T, Ji D, Wang P, et al. The atypical protein kinase RIOK3 contributes to glioma cell proliferation/survival, migration/invasion and the AKT/mTOR signaling pathway. Cancer Lett. 2018;415:151–163. doi:10.1016/j.canlet.2017.12.010
25. Gao S, Jin L, Liu G, et al. Overexpression of RASD1 inhibits glioma cell migration/invasion and inactivates the AKT/mTOR signaling pathway. Sci Rep. 2017;7(1):3202. doi:10.1038/s41598-017-03612-0
26. Iser IC, Pereira MB, Lenz G, Wink MR. The epithelial-to-mesenchymal transition-like process in glioblastoma: an updated systematic review and in silico investigation. Med Res Rev. 2017;37(2):271–313. doi:10.1002/med.21408
27. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-seq data. Nucleic Acids Res. 2014;42(Database issue):D92–D97. doi:10.1093/nar/gkt1248
28. Ma Z. Downregulation of SETD8 by miR-382 is involved in glioma progression. Pathol Res Pract. 2018;214(3):356–360. doi:10.1016/j.prp.2018.01.004
29. Tong H, Zhao K, Zhang J, Zhu J, Xiao J. YB-1 modulates the drug resistance of glioma cells by activation of MDM2/p53 pathway. Drug Des Devel Ther. 2019;13:317–326. doi:10.2147/DDDT.S185514
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.