Back to Journals » OncoTargets and Therapy » Volume 11
The role of gene sculptor microRNAs in human precancerous lesions
Authors Wang RH , He LY, Zhou SH
Received 16 April 2018
Accepted for publication 18 June 2018
Published 10 September 2018 Volume 2018:11 Pages 5667—5675
DOI https://doi.org/10.2147/OTT.S171241
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Ran-Hong Wang,1,2 Lan-Ying He,1 Shui-Hong Zhou1
1Department of Otolaryngology, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou City, People’s Republic of China; 2Department of Otolaryngology, The Third Hospital of Hangzhou City, Hangzhou City, People’s Republic of China
Abstract: MicroRNAs (miRNAs) are small noncoding RNAs. These noncoding RNAs regulate the expression of target genes and inhibit the translation of target proteins at the post-transcriptional level. miRNAs also play an important role in human health, from the development and differentiation of cells to the occurrence and progression of disorders such as cancer, cardiovascular diseases, and neurodegenerative diseases. Precancerous lesions are lesions prior to invasive carcinomas, and carcinogenesis is a very complicated process, which is multistage and the result of multigene synergy. miRNAs exert effects as both oncogenes and tumor suppressor genes by regulating target genes involved in signaling pathways. Hence, precancerous lesions are accompanied by relevant miRNA changes. Based on the morphology of miRNAs in vivo and the specificity of miRNA, various novel miRNA analysis methods have been developed, including reverse transcription quantitative PCR, enzyme analysis, molecular beacons, and deep sequencing. For example, in the laryngeal epithelial precancerous lesions, the data demonstrate that the expression of miR-10a-5p is downregulated and miR-484 is the most abundant miRNA in hepatic precancerous lesions. In this review, we discuss the functional roles of miRNAs in human precancerous lesions.
Keywords: miRNAs, precancerous lesions, therapy
Introduction
MicroRNAs (miRNAs)
miRNAs are small noncoding RNAs, which have been shown recently to contain ~21–25 nucleotides. The miRNAs are widely found in mammals, nematodes, drosophila, and plants. These noncoding RNAs regulate the expression of target genes and inhibit the translation of target proteins at the post-transcriptional level. miRNAs also play an important role in human health, from the development and differentiation of cells to the occurrence and progression of disorders such as cancer, cardiovascular diseases, and neurodegenerative diseases. Multiple studies have shown that miRNAs exert effects as both oncogenes and tumor suppressor genes by regulating target genes involved in signaling pathways, key miRNAs regulate the expression levels of hundreds of genes simultaneously, and other many types of miRNAs regulate their targets cooperatively;1 the miRNA milieu, unique to each cell type, provides important context for the evolution of all mRNA sequences and is productively used to dampen the utilization of thousands of mRNAs.2 Transcription and other nuclear events set up a column of gene expression, and then, the miRNAs, as a stone sculptor, play an important role in multiple physiological and pathological functions.3 Hence, elucidation of the functional roles of miRNAs in human precancerous lesions could assist in the prevention and therapy of cancer.
The generation and regulatory mechanisms of miRNAs
The human genome encodes ~3,000 unique miRNAs, with the majority being specific to humans, accounting for ~1% of the total human genome.4 However, miRNAs are not transcribed from genes directly. Instead, longer primary miRNAs (pri-miRNAs) are transcribed from the corresponding genes by RNA polymerase II. The pri-miRNAs are cleaved into miRNA precursors (pre-miRNAs) with hairpin-like structures containing ~70 nucleotides by a protein complex formed by Drosha and DGCR8 of the RNase III family within nuclei, followed by transfer into the cytoplasm by Exportin-5/Ran-GTP. Ultimately, pre-miRNAs are cleaved into mature miRNAs containing ~22 nucleotides by Dicer, another member of the RNase III family, subsequently forming an RNA-induced silencing complex, which binds to the 3′-UTR of a target gene mRNA to regulate its expression.5 Early annotation of miRNAs was based on their sequences and used a numbering system to classify individual miRNA sequences, which are highly conserved across species, with the same RNA sequences found among many mammalian species including humans (Figure 1).6
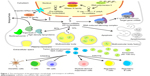  | Figure 1 The mechanisms of the generation, morphology, and transport of miRNAs. |
As an important regulatory molecule of in vivo gene expression, each miRNA can regulate hundreds of target genes and different miRNAs can coordinately regulate the same mRNA. miRNAs and their target molecules together form a complicated regulatory network, which controls multiple activities in cells.7 The abnormal expression of miRNAs results in a series of molecular changes. For example, modifications of the promoter region can alter the expression patterns. Hypermethylation of CpG islands and acetylation in promoter regions have been confirmed to reduce and increase miRNA expressions, respectively, and the miRNA promoter itself is also restricted by some transcription factors, thereby playing a regulatory role. It has been reported that the intracellular expression levels of miRNAs harboring oncogenic functions (eg, miR-106-25, miR-27a, miR-221/222, and miR-7-9) are elevated, which subsequently promote G1/S conversion and accelerate cell cycle progression. In contrast, the overexpression of miRNAs exhibiting antitumor effects (eg, the let-7 family, miR-15/16, miR-34 family, miR-124/137, miR-107, and miR-19) may contribute to G0/G1 cell cycle arrest and prevent cell cycle progression.8
The biological functions of miRNAs
miRNAs are involved mainly in the regulation of gene expression. Gene expression, including transcription and translation, is a strictly controlled and complicated process. Post-transcriptional modification of the 3′-UTR is a crucial step that determines the rate of translation from mRNA to protein.9 miRNAs also play important roles in multiple physiological and pathological functions, including development, cell proliferation, hematopoiesis, and cell death. Increasing studies have shown that tissue repair and injury, immune and inflammatory reactions, and metabolism are also largely regulated by miRNAs.10
The role of miRNAs in angiogenesis
Germination of blood vessels guarantees an adequate blood supply for cancer cells. In addition, even more blood vessels are necessary for tumor growth and metastasis. The role of miRNAs in the regulation of angiogenesis involves vascular miRNAs.11 For example, miR-126, found in exosomes derived from bone marrow suppressor cells, has been identified as a vascular miRNA associated with pulmonary metastasis in mouse breast cancer 4T1 cells.12 Cytokine VEGF is a proangiogenic factor, and the transcription factor HIF-1α is an antiangiogenic factor, both of which are directly or indirectly controlled by miRNAs during angiogenesis.13 miR-210 is secreted and expressed in lung adenocarcinoma cells induced by the activation of the TIMP-1 pathway, which downregulates the antiangiogenic gene HIF-1α, thereby promoting angiogenesis.14 In contrast, miR-16 released by mesenchymal stem cells in the tumor microenvironment reduces angiogenesis by inhibiting VEGF.15 In nontransformed human bronchial epithelial cells cultured from transformed human bronchial epithelial cells, endothelial formation occurs in the presence of elevated miR-21, which is a marker of angiogenesis.16
The role of miRNAs in immunity
The expression and roles of miRNAs can be illustrated by the regulation of innate and adaptive immune cells. Immune cells consist of innate immune cells, including neutrophils, macrophages, dendritic cells (DCs), mast cells, myeloid-derived suppressor cells, and natural killer (NK) cells, as well as adaptive immune cells, including T lymphocytes, B lymphocytes, and NK cells. Among these cells, NK cells are a subset of lymphocytes that rapidly respond to the presence of tumor cells and trigger antitumor immune reactions.17 miRNA provided the best-characterized processes for immune cell development, and genetic deletion of miR-223 results in an increased expression of its target myocyte-specific enhancer factor 2C (MEF2C), which drives myeloid progenitor proliferation and expansion of the granulocyte lineage. The zinc finger protein GFI1 is a transcriptional repressor that is crucial for granulocyte differentiation and directly binds to the miR-21 and miR-196b promoters; enforced expression of these two miRNAs results in a block in granulocyte differentiation.18 Immune cells generate various growth factors, angiogenic factors, proteases, chemokines, and cytokines, thereby resulting in the clearance of tumor cells or formation of immunosuppressive microenvironments. Relevant immune regulation of miRNAs can serve as a bridge between immune reactions and cancer.19 miR-181a can regulate B-cell differentiation and T-cell differentiation, and a lack of miR-155 leads to B-cell immunodeficiency. The miR-17-92 series is detected in T cells and DCs, and miR-34c and miR-214 are considered damage-associated molecules.20 In macrophages, miR-125b, miR-146, and miR-155 function as pathogenicity-associated molecules. Macrophages exist as two distinct phases in the tumor microenvironment, such as antitumorigenic M1 macrophages and tumorigenic M2 macrophages, and the polarization of M1 to M2 macrophages is regulated by miRNAs.21 Intriguingly, miRNAs target not only cancer cells but also stromal cells, such as cancer-associated fibroblasts and lymphocytes, thereby transforming the normal matrix into tumor-associated stroma, which is critical to carcinogenesis as well as tumor progression and metastasis.22
The role of miRNAs in metabolism
miR-22, let-7a, and miR-125b prevent mitochondrial oxidative phosphorylation and induce metabolic changes in tumor cells, mimicking a hypoxia-induced phenotype.23 The miRNAs miR-22-3p, let-7a-5p, and miR-125b-5p are secreted by cancer-associated fibroblasts and promote the viability of prostate cancer cells by targeting oxidative phosphorylation-associated genes, such as cytochrome C oxidase I and cytochrome B.24 The miR-122 levels are elevated in extracellular vesicles secreted by breast cancer cells. These cells are absorbed by noncancerous brain astrocytes and lung fibroblasts, the areas of which are initiated as “pre-metastatic niches.” In these noncancerous cells, elevated levels of miR-122 reduce the levels of glycolytic enzymes, pyruvate kinase, and glucose transporter 1 in the brain astrocytes and lung fibroblasts, which allow cancer cells to regulate their supply of glucose and ATP production.25 miR-122 is the most abundantly expressed miRNA in hepatocytes, and its functions include the regulation of lipid metabolism, replication of hepatitis C virus, cell differentiation, and hepatic metabolism. Reduced expression of miR-122 leads to mitochondrial dysfunction and subsequently hepatic dysfunction.26 A recent study has reported that miR-126, considered a vascular miRNA, also controls tumor metabolism by suppressing insulin receptor substrate 1.27 Insulin receptor substrate 1 is an insulin-like growth factor 1 receptor or scaffolding protein of the insulin receptor, which can enhance the signals of these receptor pathways.28 Cancer cells and secretory miRNAs generated by cells in the tumor microenvironment regulate metabolic pathways and promote survival and metastasis in autocrine, paracrine, and endocrine patterns.29
The role of miRNA in inflammation
Inflammation is initiated by innate immune cells in response to external stimuli such as pathogen-associated molecular patterns and host-derived damage-associated molecular patterns. These cells initiate signaling cascades that activate key transcription factors and regulators such as NF-κB, AP1, and MAPKs, all of which regulate inflammation-specific genes. NF-κB signaling is affected by miRNAs, which target either the upstream NF-κB activating kinases (IKKα or IKKβ) or other NF-κB signaling components.30 In prostate cancer, the tumor suppressive miR-497 regulates NF-κB signaling by targeting IKKβ, which activates canonical NF-κB signaling leading to the inhibition of prostate cancer cell proliferation, migration, and invasion. Importantly, miR-497 expression is reduced in prostate cancer cells, leading to a more aggressive tumor phenotype.31 miR-520/373 was further analyzed and was shown to inhibit NF-κB in estrogen-negative breast cancer cells, which further resulted in the downregulation of NF-κB targets such as the proinflammatory cytokines IL-6, IL-8, CXCL1, and ICAM-1, leading to the inhibition of tumor-related inflammation and suppression of tumor growth and metastasis. miR-199a negatively regulates the expression of IKKβ in ovarian cancer cells and inhibits the secretion of proinflammatory cytokines, thereby causing the suppression of tumor progression and chemoresistance.32 NF-κB also influences the expression of miRNAs. Several miRNAs, including miR-9, miR-21, miR-30b, miR-143/miR-145, miR-146a, miR-155, miR-221/222, miR-224, miR-301a, and the miR-17-92 clusters, have been validated as targets of the NF-κB transcription factors, In tumor-associated inflammation, the proinflammatory cytokine IL-1 leads to the activation of NF-κB and subsequent upregulation of miR-425 in gastric cancer cells. miR-140 acts as a liver tumor suppressor by negatively regulating NF-κB activity by directly targeting DNA methyltransferase 1 (Dnmt1) expression. In this cellular context, NF-κB suppresses miR-140 expression, resulting in the upregulation of DNMT1 and increased NF-κB activity, forming a positive feedback loop that promotes liver cancer. Finally, an interesting example of NF-κB-regulated miRNAs is that of miR-221/222, a miRNA family with a dual functional role, acting, in different cellular contexts, either as oncomiRs promoting cancer progressionor as tumor suppressors, promoting cellular senescence.33
The morphology and transport of miRNAs in vivo
It is now widely accepted that ~10% of miRNAs are secreted via exosomes. The remaining 90% are transported in stable complexes such as Argonaute 2, high-density lipoproteins, and nucleophosmin 1. These complexes may protect miRNAs from degradation in body fluids. Circulatory miRNAs in bodily fluids are either complexed with proteins or packaged in exosomes and called “secretory miRNAs”.34 Exosomes are lipoprotein complexes 40–100 nm in diameter containing endocytosis and are formed by internal budding of the endometrial membrane. They are intracellular multivesicular bodies that subsequently fuse with the plasma membrane, followed by their release into the extracellular space. The miRNA-containing complexes are released into biological fluids, such as blood, saliva, milk, urine, and cerebrospinal fluid. These complexes serve as small RNA delivery systems, which likely interact with distant cells via blood circulation, allowing miRNA-mediated cellular interactions.35 Small, closed membrane vesicles generated during apoptosis, called apoptosomes, can deliver specific miRNAs.36 The delivered miRNAs play a role in recipient cells where they regulate target genes and their corresponding signaling pathways. Tumor-derived exosomes can promote immune escape of cancer cells through direct interactions with immune cells, inhibiting T cells, DCs, and NK cells and inducing immunosuppressive cells, such as myeloid derived suppressor cells and regulatory T and B cells.37 These miRNAs are considered to be “dark matter”, which exists almost everywhere around us but almost never escapes from all our sight. This discovery is an important complement to the secondary, intermediary role of RNA in central metabolism.
Detection methods
The isolation of sterile secretory miRNAs is a challenge in studying the physiological significance of these miRNAs in bodily fluids. Pure exosomes must be isolated prior to RNA extraction. Exosome-associated structural features, including density, size, shape, and surface markers, are used in exosome isolation techniques. The currently available approaches include ultracentrifugation, immunoaffinity capture, and isolation techniques. Ultracentrifugation is considered the “gold standard” for isolating exosomes.38 In ultracentrifugation, exosome enrichment is based on size, mass, and density to remove contaminants, including intact cells and cellular debris. Exosomes can be compressed into balls, allowing density gradient centrifugation to be used for purification.39 Size-based isolation techniques have also been used to isolate exosomes, in which particles are separated by molecular mass or size exclusion chromatography. Although this method is faster than ultracentrifugation, the filtration forces may disrupt larger vesicles, and hence proper control is necessary.40 Adherent exosomes can also be isolated by immunoaffinity capture techniques using submicron-sized magnetic particles targeting the surface proteins expressed on exosomes.41
After obtaining high-purity exosomes, miRNAs can be extracted from the vesicles.42 Reverse-transcription quantitative polymerase chain reaction is a traditional and routine method for detecting miRNAs in real time and has the significant advantages of a high sensitivity, wide dynamic range, and great precision.43 However, this technique is hampered by the short length of miRNAs (21–25 nucleotides) and the similarity among homologous miRNA family sequences.
Various novel miRNA analysis methods incorporating bioluminescence, enzymatic analyses, molecular beacons, deep sequencing, and chain reactions based on lock nucleic acids have been developed but with certain unavoidable drawbacks. The bioluminescent protein Renilla luciferase is used as a marker in bioluminescence assays using a simple operation.44 Enzymatic analysis has the advantage of strong signals but the disadvantages of relatively low specificity and sensitivity.45 Deep sequencing can be used to analyze absolute miRNA levels rapidly and quantitatively but at a relatively high cost.46 Chain reactions based on lock-nucleic acids require immobilization47 and separation as well as the design of complicated DNA probes,48 which limit its application. Because of the short length and low abundance of miRNAs, it is necessary to amplify the signal to improve the sensitivity of detection. It has been reported that single-molecule array (Simoa) technology uses a sandwiched ultrasensitive detection method to detect multiple miRNAs directly without preamplification, which is effective for a range of different target sequences and is a generic method for miRNA detection.49 However, multiple experimental challenges, such as standardized methods and strategies, remain to be solved for each of the above detection methods.
miRNAs and precancerous lesions
Precancerous lesions
Medically, the general order of lesion progression is “early biological effects,” followed by “early disease”, eventually “disease states,” and ultimately cancer as the final stage of disease progression.50 Precancerous lesions occur prior to invasive carcinomas, which are defined as any morphologically differentiable, proliferative lesion (atypical cell differentiation).51 However, in clinical practice, the definition of a precancerous lesion is not clear, because it is not cancerous and exhibits variations at the molecular and cellular levels in the pathway leading to cancer. Therefore, precancerous lesions should contain the following characteristics: 1) association with an increased risk of cancer, which excludes cellular atypia, monoclones, cytogenetic marker mutations, and specific mutations; 2) cancer cells originate from the precancerous lesion cells upon progression to cancer; 3) the precancerous tissue differs from normal tissue; 4) precancerous lesions differ from tumor progression despite certain (not all) molecular and phenotypic features of cancerous features; and 5) a clear diagnosis of precancerous lesions.52 Under most circumstances, the precancerous lesions include dysplasia, atypical hyperplasia, carcinoma in situ, epithelial sarcoma, and early malignancy. However, in practice, the most commonly used distinctions between precancerous lesions and cancer include matrix infiltration and diffusion potential.53 Many epithelial tumors tend to extend into adjacent normal glands or tubular structures and mimic intrusions. During the progression from precancerous lesions to invasion into stroma, cancer cells exhibit self-sufficient growth, insensitivity to antigrowth signals, escape from apoptosis, unlimited replication potential, and sustained angiogenesis, as well as tissue infiltration and metastasis of certain malignant cells, although the majority of cells in the lesion are still noninvasive.54 Most cancers have identifiable precancerous lesions. For example, cervical intraepithelial neoplasia progresses to cervical squamous cell carcinoma, bronchial squamous epithelial hyperplasia progresses to lung squamous cell carcinoma, colorectal adenoma progresses to colorectal cancer, and vocal leukoplakia progresses to laryngeal cancer. These precancerous lesions have common causes, including long-term smoking, infections, bronchial epithelial metaplasia, and stimulation of gastroesophageal reflux.
Mechanisms of carcinogenesis
The mechanism of carcinogenesis is a very complicated process involving multiple stages and multiple gene synergies (Figure 2). Carcinogenesis results from the activation of various oncogenes and the inactivation (or deletion) of tumor suppressor genes, including gene mutations, gene amplifications, chromosomal translocations, and gene insertions. Mutations of oncogenes and tumor suppressor genes include both germline and somatic mutations. The majority of tumor genetic changes are acquired (somatic mutations). Activation of oncogenes and inactivation of tumor suppressor genes eventually lead to uncontrolled cell division and ultimately to tumorigenesis.55 The products of oncogenes and tumor suppressor genes are involved in many signaling systems and levels, from the outside of the cell membrane to the nucleus, leading to an imbalance between cell growth and differentiation and eventually metastatic cancer by regulating extracellular and membrane signal transductions, G proteins and GTPase activating proteins, signal transmission from the membrane to the cytoplasm, intercellular signal transmission, signal transmission from the cytoplasm to the nucleus, and intranuclear signal transmission.56 The abnormal expression of miRNAs in human tissues and blood is closely associated with the occurrence, progression, and therapeutic response of precancerous lesions. Hence, miRNAs are very promising as targets for cancer therapeutic agents.
  | Figure 2 The role of miRNAs in different aspects of carcinogenesis. |
Specific miRNA expression in tumors
Based on the molecular mechanism of miRNA, does this mean that many of these other mRNAs would in fact be targeted if expressed in the same cells as the cognate miRNAs? Perhaps not the miRNA milieu, unique to each cell type, provides important context for the evolution of all mRNA sequences and is productively used to dampen the utilization of thousands of mRNAs. For mRNAs that should not be expressed in a particular cell type, miRNAs reduce protein production to inconsequential levels. miRNAs could adjust protein output in a manner that allows for customized expression in different cell types yet a more uniform level within each cell type. The quality and stability of base pairing are in fact the primary determinant of specificity – should also be considered. After all, this complementarity requirement includes a seven-nt perfect or near-perfect core match near the 5′ terminus of the miRNA,57 which by itself would represent a degree of specificity comparable to that of the DNA sites recognized by many transcription factors. Pairing outside the 7 nt core site, although perhaps less important than once thought, provides means of conferring added specificity,58 and the abnormal expression of specific miRNAs can be detected in precancerous lesions and tumors at the molecular and cellular levels.
Compared with healthy controls, certain miRNAs are upregulated in patients with colorectal cancer, even during the early stages, compared with seven in vitro cells (let-7a, miR-21, miR-98, miR-146, and miR-183).59 In pancreatic tumors, miR-20a, miR-21, miR-24, miR-25, miR-26b, and miR-99a are significantly upregulated,60 while miR-126, miR-145, miR-223, miR-492, miR-505, and miR-663a are downregulated.61 In breast tumors, miR-96-5p, miR-127-3p, miR-133a, miR-145, and miR-148a are upregulated,62 while miR-103, miR-107, miR-195, miR-200c, and miR-451 are significantly downregulated.63 In prostate cancer, miR-326, miR-328, miR-331-3p, and miR-375 are upregulated,64 while miR-30c, miR-181a-2, let-7a, let-7c, let-7e, and miR-432 are downregulated.65 In melanoma, miR-210 and miR-211 are upregulated,66 while miR-29c, miR-324-3p, miR-509-3p, and miR-509-5p are downregulated.67 In bladder cancer, miR-26b-5p, miR-144-5p, miR-148b-3p, and miR-152 are upregulated,68 while miR-15b-5p, miR-27a-3p, miR-30a-5p, and miR-92a are downregulated.69 miRNAs circulating in blood identified as candidate biomarkers of various cancers are provided in Table 1. Those miRNAs downregulated in tumor tissues are called tumor suppressor miRNAs. Therefore, precancerous lesions are accompanied by relevant changes in miRNA expression.70 For example, in laryngeal epithelial precancerous lesions, the expression of miR-10a-5p is downregulated while that of miR-34c-5p is upregulated; miR-10a-5p expression is positively correlated with the grade of laryngeal epithelial precancerous lesions and sex of the patient, while miR-34c-5p expression is associated with alcohol consumption. Statistical analyses have shown that miR-10a expression in laryngeal epithelial precancerous lesions is higher in female patients than in male patients. In contrast, the expression of miR-34c is lower in female than male patients and the expression of miR-34c is higher in drinkers than in nondrinkers.71 miR-484 is the most abundant miRNA in hepatic precancerous lesions. Bioinformatic and functional studies have shown that miR-484 regulates the transforming growth factor beta and nuclear factor-κB pathways to induce hepatocellular carcinogenesis, while depletion of miR-484 in mice prevented precancerous lesion progression and tumorigenesis.72 Intraepithelial neoplasia is considered a precancerous lesion of esophageal squamous cell carcinoma. E-cadherin, encoded by CDH1, is an important suppressor of tumor metastasis and a potential target of miR-25. Low and high E-cadherin levels are observed in cell lines with high and low miR-25 expressions, respectively. In addition, miR-25 can promote cell proliferation by inhibiting E-cadherin. Hence, miR-25 expression may be elevated in in esophageal squamous cells in intraepithelial neoplasia compared with normal tissue.73
  | Table 1 List of miRNAs deregulated in human neoplasm and precancerous lesions |
Conclusion and prospects
The identification of specific miRNAs in sample tissues or biological fluids would be critical not only for understanding the physiological and pathological processes of diseases but also for treating cancerous cells during the early stages by taking advantage of miRNA mechanisms. Overall, these future studies have the potential to assist the development of novel therapeutic strategies for cancer treatment.
Acknowledgments
This work was supported by Science and Technology Department of Zhejiang Province, People’s Republic of China (no 2016C33144). The English in this document has been checked by at least two professional editors, both native speakers of English (for a certificate, refer http://www.textcheck.com/certificate/VYqSUq).
Author contributions
R-HW designed and wrote the article. L-YH collected the materials and reviewed the literatures. S-HZ revised the article. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–874. | ||
Bartel DP. MicroRNAs: genomics. biogenesis, mechanism, and function. Cell. 2004;116:281–297. | ||
Bartel DP. Metazoan microRNAs. Cell. 2018;173(1):20–51. | ||
Londin E, Loher P, Telonis AG, et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc Natl Acad Sci U S A. 2015;112(10):E1106–E1115. | ||
Kai K, Dittmar RL, Sen S. Secretory microRNAs as biomarkers of cancer. Semin Cell Dev Biol. 2017;78:22–36. | ||
Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. | ||
Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3(11):881–886. | ||
Bueno MJ, Malumbres M. MicroRNAs and the cell cycle. Biochim Biophys Acta. 2011;1812(5):592–601. | ||
di Leva G, Croce CM. miRNA profiling of cancer. Curr Opin Genet Dev. 2013;23(1):3–11. | ||
Wang J, Chen J, Sen S. MicroRNA as biomarkers and diagnostics. J Cell Physiol. 2016;231(1):25–30. | ||
Li J, Zhang Y, Liu Y, et al. Microvesicle-mediated transfer of microRNA-150 from monocytes to endothelial cells promotes angiogenesis. J Biol Chem. 2013;288(32):23586–23596. | ||
Taverna S, Amodeo V, Saieva L, et al. Exosomal shuttling of miR-126 in endothelial cells modulates adhesive and migratory abilities of chronic myelogenous leukemia cells. Mol Cancer. 2014;13:169. | ||
Umezu T, Tadokoro H, Azuma K, Yoshizawa S, Ohyashiki K, Ohyashiki JH. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood. 2014;124(25):3748–3757. | ||
Cui H, Seubert B, Stahl E, et al. Tissue inhibitor of metalloproteinases-1 induces a pro-tumourigenic increase of miR-210 in lung adenocarcinoma cells and their exosomes. Oncogene. 2015;34(28):3640–3650. | ||
Lee JK, Park SR, Jung BK, et al. Exosomes derived from mesenchymal stem cells suppress angiogenesis by down-regulating VEGF expression in breast cancer cells. PLoS One. 2013;8(12):e84256. | ||
Liu Y, Luo F, Wang B, et al. STAT3-regulated exosomal miR-21 promotes angiogenesis and is involved in neoplastic processes of transformed human bronchial epithelial cells. Cancer Lett. 2016;370(1):125–135. | ||
Cao W, Cheng W, Wu W. MicroRNAs reprogram tumor immune response. Methods Mol Biol. 2018;1699:67–74. | ||
Mehta A, Baltimore D. MicroRNAs as regulatory elements in immune system logic. Nat Rev Immunol. 2016;16(5):279–294. | ||
Paladini L, Fabris L, Bottai G, Raschioni C, Calin GA, Santarpia L. Targeting microRNAs as key modulators of tumor immune response. J Exp Clin Cancer Res. 2016;35:103. | ||
Sonkoly E, Ståhle M, Pivarcsi A. MicroRNAs and immunity: novel players in the regulation of normal immune function and inflammation. Semin Cancer Biol. 2008;18(2):131–140. | ||
Roy S. miRNA in macrophage development and function. Antioxid Redox Signal. 2016;25(15):795–804. | ||
Negrini M, Ferracin M, Sabbioni S, Croce CM. MicroRNAs in human cancer: from research to therapy. J Cell Sci. 2007;120(pt 11):1833–1840. | ||
vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. | ||
Zhao H, Yang L, Baddour J, et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. Elife. 2016;5:e10250. | ||
Fong MY, Zhou W, Liu L, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17(2):183–194. | ||
Hsu SH, Wang B, Kota J, et al. Essential metabolic, anti-inflammatory, and anti-tumorigenic functions of miR-122 in liver. J Clin Invest. 2012;122(8):2871–2883. | ||
Tao H, Wang MM, Zhang M, et al. MiR-126 suppresses the glucose-stimulated proliferation via IRS-2 in INS-1 β cells. PLoS One. 2016;11(2):e0149954. | ||
Choi JW, Um JH, Cho JH, Lee HJ. Tiny RNAs and their voyage via extracellular vesicles: secretion of bacterial small RNA and eukaryotic microRNA. Exp Biol Med. 2017;242(15):1475–1481. | ||
Mann M, Mehta A, Zhao JL, et al. An NF-κB-microRNA regulatory network tunes macrophage inflammatory responses. Nat Commun. 2017;8(1):851. | ||
Kong XJ, Duan LJ, Qian XQ, et al. Tumor-suppressive microRNA-497 targets IKKβ to regulate NF-κB signaling pathway in human prostate cancer cells. Am J Cancer Res. 2015;5(5):1795–1804. | ||
Keklikoglou I, Koerner C, Schmidt C, et al. MicroRNA-520/373 family functions as a tumor suppressor in estrogen receptor negative breast cancer by targeting NF-κB and TGF-β signaling pathways. Oncogene. 2012;31(37):4150–4163. | ||
Chen R, Alvero AB, Silasi DA, et al. Regulation of IKKbeta by miR-199a affects NF-kappaB activity in ovarian cancer cells. Oncogene. 2008;27(34):4712–4723. | ||
Markopoulos G, Roupakia E, Tokamani M, et al. Roles of NF-κB signaling in the regulation of miRNAs impacting on inflammation in cancer. Biomedicines. 2018;6(2):40. | ||
Thayanithy V, O’Hare P, Wong P, et al. A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication. Cell Commun Signal. 2017;15(1):46. | ||
Lv LL, Cao Y, Liu D, et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9(10):1021–1031. | ||
Kim J, Shin H, Park J. RNA in salivary extracellular vesicles as a possible tool for systemic disease diagnosis. J Dent Res. 2017;96(8):938–944. | ||
Geis-Asteggiante L, Belew AT, Clements VK, et al. Differential content of proteins, mRNAs, and miRNAs suggests that MDSC and their exosomes may mediate distinct immune suppressive functions. J Proteome Res. 2018;17(1):486–498. | ||
Ma C, Liu S, Shi C. Ultrasensitive detection of microRNAs based on hairpin fluorescence probe assisted isothermal amplification. Biosens Bioelectron. 2014;58:57–60. | ||
Eldh M, Lötvall J, Malmhäll C, Ekström K. Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Mol Immunol. 2012;50(4):278–286. | ||
Quintana JF, Makepeace BL, Babayan SA, et al. Extracellular Onchocerca-derived small RNAs in host nodules and blood. Parasit Vectors. 2015;8:58. | ||
Nakai W, Yoshida T, Diez D, et al. A novel affinity-based method for the isolation of highly purified extracellular vesicles. Sci Rep. 2016;6:33935. | ||
Jia YJ, Zhou ML, Exosomes ZSH. Microvesicles, and head and neck cancers. Int J Clin Exp Med. 2016;9:15040–15049. | ||
Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10(10):704–714. | ||
Cissell KA, Campbell S, Deo SK. Rapid, single-step nucleic acid detection. Anal Bioanal Chem. 2008;391(7):2577–2581. | ||
Allawi HT, Dahlberg JE, Olson S, et al. Quantitation of microRNAs using a modified Invader assay. RNA. 2004;10(7):1153–1161. | ||
Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief Bioinform. 2009;10(5):490–497. | ||
Lee JM, Jung Y. Two-temperature hybridization for microarray detection of label-free microRNAs with attomole detection and superior specificity. Angew Chem Int Ed Engl. 2011;50(52):12487–12490. | ||
Zhang P, Zhang J, Wang C, Liu C, Wang H, Li Z. Highly sensitive and specific multiplexed microRNA quantification using size-coded ligation chain reaction. Anal Chem. 2014;86(2):1076–1082. | ||
Cohen L, Hartman MR, Amardey-Wellington A, Walt DR. Digital direct detection of microRNAs using single molecule arrays. Nucleic Acids Res. 2017;45(14):e137. | ||
Caporaso NE. Matter Wprecursors. Why precursors matter. Cancer Epidemiol Biomarkers Prev. 2013;22(4):518–520. | ||
Pontén J. Cell biology of precancer. Eur J Cancer. 2001;37(suppl 8):97–113. | ||
Berman JJ, Albores-Saavedra J, Bostwick D, et al. Precancer: a conceptual working definition – results of a consensus conference. Cancer Detect Prev. 2006;30(5):387–394. | ||
Wacholder S. Precursors in cancer epidemiology: aligning definition and function. Cancer Epidemiol Biomarkers Prev. 2013;22(4):521–527. | ||
Ponten J. Cell biology of precancer. Eur J Cancer. 2001;37:S97–S113. | ||
Vassilev A, Depamphilis ML. Links between DNA replication, stem cells and cancer. Genes (Basel). 2017;8(2):45. | ||
Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13(6):472–482. | ||
Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1(3):E60. | ||
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. | ||
Giráldez MD, Lozano JJ, Ramírez G, et al. Circulating microRNAs as biomarkers of colorectal cancer: results from a genome-wide profiling and validation study. Clin Gastroenterol Hepatol. 2013;11688(6):681.e3–688.e3. | ||
Li A, Yu J, Kim H, et al. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin Cancer Res. 2013;19(13):3600–3610. | ||
Kojima M, Sudo H, Kawauchi J, et al. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS One. 2015;10(2):e0118220. | ||
Cuk K, Zucknick M, Madhavan D, et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS One. 2013;8(10):e76729. | ||
Zhao FL, Dou YC, Wang XF, et al. Serum microRNA-195 is down-regulated in breast cancer: a potential marker for the diagnosis of breast cancer. Mol Biol Rep. 2014;41(9):5913–5922. | ||
Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4(7):e6229. | ||
Bryant RJ, Pawlowski T, Catto JWF, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106(4):768–774. | ||
Stark MS, Klein K, Weide B, et al. The prognostic and predictive value of melanoma-related microRNAs using tissue and serum: a microRNA expression analysis. EBioMedicine. 2015;2(7):671–680. | ||
Greenberg E, Besser MJ, Ben-Ami E, et al. A comparative analysis of total serum miRNA profiles identifies novel signature that is highly indicative of metastatic melanoma: a pilot study. Biomarkers. 2013;18(6):502–508. | ||
Jiang X, du L, Wang L, et al. Serum microRNA expression signatures identified from genome-wide microRNA profiling serve as novel noninvasive biomarkers for diagnosis and recurrence of bladder cancer. Int J Cancer. 2015;136(4):854–862. | ||
du M, Shi D, Yuan L, et al. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep. 2015;5:10437. | ||
Kwan JY, Psarianos P, Bruce JP, Yip KW, Liu FF. The complexity of microRNAs in human cancer. J Radiat Res. 2016;57(suppl 1):i106–i111. | ||
Hu Y, Liu H. MicroRNA-10a-5p and microRNA-34c-5p in laryngeal epithelial premalignant lesions: differential expression and clinicopathological correlation. Eur Arch Otorhinolaryngol. 2015;272(2):391–399. | ||
Yang Y, Lin X, Lu X, et al. Interferon-microRNA signalling drives liver precancerous lesion formation and hepatocarcinogenesis. Gut. 2016;65(7):1186–1201. | ||
Xu X, Chen Z, Zhao X, et al. MicroRNA-25 promotes cell migration and invasion in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2012;421(4):640–645. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
