Back to Journals » Infection and Drug Resistance » Volume 12
The role of cfa gene in ampicillin tolerance in Shigella
Authors Liu Q , Li M, Teng Y, Yang H, Xi Y, Chen S, Duan G
Received 8 April 2019
Accepted for publication 12 August 2019
Published 5 September 2019 Volume 2019:12 Pages 2765—2774
DOI https://doi.org/10.2147/IDR.S211550
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Video abstract presented by Qiaoli Liu.
Views: 159
Qiaoli Liu, Mengchen Li, Yanli Teng, Haiyan Yang, Yuanli Xi, Shuaiyin Chen, Guangcai Duan
Department of Epidemiology and Health Statistics, College of Public Health, Zhengzhou University, Zhengzhou 450001, People’s Republic of China
Correspondence: Shuaiyin Chen; Guangcai Duan
Department of Epidemiology and Health Statistics, College of Public Health, Zhengzhou University, Zhengzhou 450001, People’s Republic of China
Tel +86 1 352 340 8394
Fax +86 6 778 1453
Email [email protected]; [email protected]
Background: Bacterial dysentery is an intestinal infectious disease caused by Shigella. The resistance of Shigella species to ampicillin has increased rapidly. Besides resistance, bacteria in a state of tolerance to antibiotics can also lead to the failure of infectious diseases therapy.
Purpose: The genetic mechanism of antibiotic tolerance remains largely unexplored. The current study aimed to investigate the mechanisms of antibiotic tolerance and to provide novel strategies for the prevention of drug resistance of Shigella.
Methods: We exposed Shigella to lethal doses of ampicillin to obtain tolerant strain. The tolerant strain was sequenced to screen non-single-nucleotide polymorphisms and Indels. We also quantitated the relative expression of gene by RT-PCR.
Results: There was one nonsynonymous mutation in the 2252304 loci of the cfa gene (G to A/Val to Met) and 10 Indels in the noncoding regions of different genes. The expression of the cfa gene was 7.56-fold higher in the tolerant strain than in the wild-type strain.
Conclusion: Our results showed that Shigella could be tolerated to ampicillin, and the cfa gene might be associated with antibiotic tolerance by increasing its expression. Our data suggest that cfa gene might be a target for overcoming drug tolerance, delaying the occurrence of drug resistance to some extent.
Keywords: Shigella, tolerance, cfa, gene mutation
Introduction
Bacillary dysentery is a prevailing acute intestinal infectious disease characterized by Shigella bacteria invading colon mucosa and inflammatory of epithelial cells.1 Approximately 163 million cases of shigellosis were reported annually, particularly in developing countries. In China, its incidence rate accounts for the top three infectious diseases and is still the prevention and control focus of public health.2 Ampicillin is a broad-spectrum antibiotic that is used to treat Enterobacteriaceae bacteria in humans as well as in animals.3 Due to the extensive abuse of ampicillin, the resistance of Shigella species to ampicillin has increased rapidly, making it more difficult to control bacillary dysentery.3
Besides resistance, bacteria in a state of tolerance to certain antibiotics can also lead to the failure of infectious disease therapy.4 Tolerance can be defined as bacteria that grow slowly or even do not grow after short exposure to high concentrations of antibiotics, and then grow again after antibiotics are discontinued.5 However, unlike resistance, the tolerant bacteria can have the same minimum inhibitory concentration (MIC); thus, it is not informative as a metric to evaluate tolerance.6 In recent years, the minimum duration of killing (MDK) is used to evaluate tolerance,7 for example, the minimum duration of treatment that is required to kill 99% of bacteria (MDK99). Till now, the mechanism of antibiotic tolerance is quite complicated and remains largely unclear. Changes in environmental conditions of bacteria often trigger the production of stress-response genes. Studies have shown that antibiotic tolerance may be caused by stress-response genes controlled by rpoS.8 Second messenger guanosine tetraphosphate (ppGpp) can lead to antibiotic tolerance.9 In addition, Aaron et al have shown that strains are isolated from patients with cystic fibrosis patients, which are biofilm bacteria that are tolerant to antibiotics, compared to plankton.10
The genetic mechanism of antibiotic tolerance has not been elucidated. To explore the mechanisms of antibiotic tolerance, in our study, we exposed Shigella to high concentrations of ampicillin to obtain tolerant strain. The tolerant strain was sequenced to screen non-single-nucleotide polymorphisms (SNPs) and Indels. Our data suggest that the cfa gene may be associated with antibiotic tolerance by increasing its expression and it might be a target for overcoming drug tolerance.
Materials and methods
Isolation and identification of bacteria
To study antibiotic tolerance of Shigella strain sensitive to ampicillin, we separated a strain sensitive to ampicillin that was originally isolated from Sui County in Henan province in 2014. This strain was confirmed by biochemical and serological identification. Biochemical identification was performed using API20E (API Systems, SA, Vercieu, France). Briefly, the strain was inoculated into Luria-Bertani (LB) agar using a sterile-inoculating loop and incubated at 37°C for 18 hrs. The colonies were picked up with sterile cotton and resuspended in 0.85% NaCl medium to achieve uniform bacterial turbidity, and the turbidity of the bacterial suspension was adjusted to 0.5 McFarland. According to the instructions of the identification manual, the bacterial resuspension was added to the tube of the test strip using a straw. The test strips of citrate utilization(CIT), acetoin production(VP), and gelatinase (GEL) should be filled with bacterial suspension.The test strips of arginine dihydrolase(ADH), lysine decarboxylase (LDC),ornithine decarboxylase (ODC),H2S production(H2S),urease(URE)should be filled with mineral oil to form an anaerobic environment. Sterile distilled water was added to the culture box, and the test strip was placed in a culture box and placed in a 37°C incubator for 18 hrs. The tryptophan deaminase (TDA) test needed to add 1 drop of TDA reagent. The indole production (IND) test needed to add a drop of JAMES reagent. The VP test needed to add a drop of VP1 and VP2. The test results were observed after 10 mins. The test results were interpreted according to the interpretation table, and the results were numerically coded, and the numerical code was found in the identification manual of API20E to determine the type of the strains.
Serological identification was completed using a slide agglutination method. In brief, a clean slide was taken, and a drop of Shigella diagnostic serum was added, and a drop of physiological saline was added as a negative control. Fresh colonies were picked and mixed thoroughly with them. They were allowed to be placed at room temperature for a while, and the flocculation or sand-like agglutination is visible to the naked eye, which is a positive reaction, and vice versa. The results were interpreted according to the test reaction. Strain sensitive to ampicillin was isolated from 39 strains by Kirby–Bauer (K-B) disk diffusion method, as recommended by the Clinical and Laboratory Standards Institute (CLSI, 2017).11 The laboratory number was mel-ss2014011. E. coli ATCC 25922 was used as a control for quality assurance.
MIC analysis of ampicillin
In order to obtain the induced concentration of Shigella on ampicillin, we measured the MIC of Shigella to ampicillin by using broth microdilution method, as recommended by the CLSI (2017).11 A bacteria was suspended in Mueller–Hinton broth (0.5 McFarland), then diluted 1000 times, aliquoted into 96-well plates and incubated at 37°C for 16 hrs; the MIC was defined as the lowest concentration of antibiotic at which no visible bacterial growth was observed. Quality control testing was performed using E. coli ATCC 25922.
Construction of ampicillin-induced tolerance model
To acquire a strain tolerant to ampicillin, we exposed Shigella strain to the lethal dose of ampicillin, which is 10 times greater than the MIC. In each cycle, a single colony was picked and cultured in Luria-Bertani (LB) broth medium, and then bacterial solution was shaken for 18 hrs to reach the stationary-growth phase at 37°C, 200 rpm, followed by 5 hrs of treatment with 10 MIC of ampicillin at 200 rpm at 37°C. The culture was washed to remove the antibiotic, resuspended in a fresh medium and grown overnight at 200 rpm at 37°C. The nonantibiotic control strains were also grown overnight for 18 hrs in Luria-Bertani (LB)broth medium, but they were not treated with any antibiotic for 5 hrs, then reinoculated in new medium. After 20 cycles of induction for both wild-type strain and nonantibiotic control strains, we stopped inducing bacteria. The induced strain had the same MIC compared with the wild-type strain, but compared with the wild-type strain, the MDK99 increased after induction, indicating that the bacteria had been tolerant.12
Determination of MDK99
To measure the survival under antibiotic treatments, the wide-type strain and tolerant strain and nonantibiotic control strain were inoculated in the Luria-Bertani (LB) broth medium, grown overnight and diluted 1:100 in fresh medium supplement with 10 MIC antibiotic. At the indicated time points, aliquots of the cultures were sampled, diluted to the appropriate dilutions and plated on Luria-Bertani (LB) agar plates. Colony forming units (CFU) were counted after incubation 24 hrs at 37°C to make sure that all CFUs had appeared. All measurements were repeated in at least two independent experiments.
Homology identification before and after induction
In order to ensure the original source of the tolerant strain, the tolerant strain was identified using 16SrRNA, biochemistry identification. Genetic DNAs from these bacteria were isolated by Bacterial Genomic DNA Kit (Shanghai Lifefeng Biotech, Shanghai, China) and sequenced by 3730XL system at the Beijing Genomics Institute (Shenzhen, China), and then, compared with the BLAST database (http://www.ncbi.nlm.nih.gov/BLAST/).13 Biochemical identification was performed using API20E (Same as above).
Sequencing and genomic analysis
To explore the genetic mechanisms of antibiotic tolerance, we extracted bacterial DNA and sequenced it for screening non-SNPs and Indels. DNA was extracted using Bacterial Genomic DNA Kit (Shanghai Lifefeng Biotech). The genome of the wide-type strain was sequenced using a PacBio RSII platform and Illumina HiSeq 4000 system (Illumina, San Diego, CA, USA) and the tolerant strain was sequenced using an Illumina HiSeq 4000 system (Illumina) at the Beijing Genomics Institute (Shenzhen, China). With alignment software MUMmer, each query sequence is aligned with the reference sequence. The variation sites between the query sequence and reference sequence are found out and filtered preliminarily to detect potential non-SNPs and Indel sites.
RNA extraction and RT-PCR
To detect the gene expression levels, RNA was extracted and RT-PCR was performed. Bacteria were inoculated in a fresh Luria-Bertani (LB) broth medium and grown to mid-logarithmic phase. Total RNA was extracted using RNA pure Bacteria kit (Beijing Com Win Biotech, Beijing, China). PrimeScriptTM RT reagent Kit (Takara Bio, Beijing, china) was used to eliminate contaminating genomic DNA according to the manufacturer’s instruction. The purified RNA was retrieved into the cDNA for the above kit. The PCR reactions (20µl total volume) contained 10 µL TB GreenTM Premix EX TapTM II, 6 µL RNase-free water, upstream and downstream primers were 0.8 µL, cDNA template 2 µL, ROX reference Dye II (50x) 0.4 µL. The PCR condition was as follows: an initial denaturation at 95°C for 30 s, 40 cycles of 5 s at 95°C and 34 s at 60°C. 2−△△CT method was used to evaluate the relative expression of genes.16SrRNA gene expression was used as an endogenous control. All data on gene expression were the mean of triplicate analysis. Gene expression was analyzed using SPSS 21.0. Two independent sample t-test was used for comparison of gene expression.
Results
Isolation and identification of strain
To study antibiotic tolerance of Shigella strain sensitive to ampicillin, we separated a strain sensitive to ampicillin that was originally isolated from Sui county in Henan province in 2014. This strain was confirmed to be Shigella by biochemical and serological identification. This Shigella strain sensitive to ampicillin was determined by K-B paper diffusion method (data not shown). As shown in Table 1, GLU, MAN and ARA could be decomposed completely (Table 1). After verification by apiweb TMM software, it was identified as Shigella.spp. Serological identification showed that it was Shigella sonnei, and we named this strain as mel-ss2014011. These results suggested that Shigella strain sensitive to ampicillin was successfully isolated.
 |
Table 1 Biochemical reaction results between wild-type strain and tolerant strain |
Induction of tolerance to ampicillin by Shigella
We tested whether the long-term treatment with ampicillin would develop a Shigella strain that was tolerant to ampicillin. To this end, wild-type strain was exposed to a Luria-Bertani (LB) broth medium for 18 hrs to reach the stationary-growth phase, followed by 5 hrs of treatment with 10 MIC of ampicillin. But nonantibiotic control strains were not treated with any antibiotic for 5 hrs. After 20 cycles of such treatment, bacteria could not be killed effectively and were tolerant to ampicillin, and we named tolerant strain as mel-ss2014011/20. The tolerant strain, wild-type and nonantibiotic control strain had the same MIC, which was 2 μg/mL (Figure 1). The bacterial concentration of the wild-type strain decreased from 6.5×1010 CFU/mL to 2.9×108 CFU/mL during 10 mins (MDK99 was 10 mins), and the bacterial concentration of nonantibiotic control strain decreased from 1.75×109 CFU/mL to 1.5×107 CFU/mL during 10 mins (MDK99 was 10 mins), but tolerant strain only reduced from 1.8×109 CFU/mL to 2.6×108 CFU/mL in 60 mins (Figure 2). In order to detect the MDK99 of the tolerant strain, we extended the time to 10 hrs and sampled every 2 hrs. The results showed that both wild-type strain and nonantibiotic control strain decreased from 1.2×1010 CFU/mL to 0 in 4 hrs, while the bacterial concentration of tolerant strain decreased from 2.8×109 CFU/mL to 5×107 more than 10 hrs (MDK99 exceeds 10 hrs) (Figure 2). The nonantibiotic control strains were consistent with the wild-type MIC and MDK99, indicating that Shigella tolerance would be generated under antibiotic pressure.
Homology identification before and after induction
To confirm that the tolerant strain was derived from a wild-type strain, 16SrRNA and biochemical was used to identify its homology. The 16SrRNA was identified, and the homology of the tolerant strain and wild-type strain to Shigella was 99% (data not shown). API20E was used to identify the tolerant (mel-ss2014011/20) strain as Shigella.spp. (Table 1). The results showed that tolerant strain was derived from the wild-type strain and the induction process was not contaminated.
Sequencing and genomic analysis
To elucidate the genetic basis of the Shigella tolerance for ampicillin, the wild-type strain was subjected to whole-genome sequencing, and the tolerant strain was sequenced. Whole-gene sequencing results showed that mel-ss2014011 contained a 4814885bp chromosome. The GC -content of chromosome is 51.99%, 5203 predicted protein-coding genes, 95 tRNA genes and 54 sRNA genes (Figure 3). Compared with the wild-type strain (mel-ss2014011), tolerant strain (mel-ss2014011/20) had one nonsynonymous mutation, and 10 Indels were located in the noncoding region. This nonsynonymous mutation was found at 2252304 loci of the GL002314 gene (G to A/Val to Met) in mel-ss2014011/20 (Figure 4). The annotation in the Kyoto Encyclopedia of Genes and Genomes (KEGG)database to the GL002314 gene is cfa, encoding cyclopropane-fatty-acyl-phospholipid synthase. Its Cluster of Orthologous Groups of Proteins (COG) function is classified as I, which is interpreted as lipid transport and metabolism (Figure 3). In addition, there were 10 single base insertions in the noncoding region of different genes (Table 2). The annotations for both GL000777 gene and GL004546 gene in the KEGG database are K07495, a putative transposase. Its COG function is classified as X, which is interpreted as Mobilome: prophages, transposons. The annotation for GL003859 gene is insB, encoding IS1 protein. Its COG function is classified as X, which is interpreted as Mobilome: prophages, transposons (Table 2). The remaining seven genes are not explained in the KEGG database (Table 2). These results indicate that it is possible that the cfa gene may be associated with Shigella tolerance to ampicillin.
 |
Figure 4 The comparison of amino acid sequences of cfa genes between wild-type and tolerant strains. Note: Subject represents the wild-type strain and Query represents the tolerant strain. |
 |
Table 2 Indels of non-coding region are screened by sequencing |
cfa expression analysis by real-time PCR
Next, we used RT-PCR to detect the expression of cfa gene. Our results showed that the relative expression levels of cfa genes in mel-ss2014011 and mel-ss2014011-20 were 1±0.14 and 7.56±0.90, respectively (Figure 5). The two independent sample t-test analysis showed a difference significant among the expression of wild-type and tolerant strains (t=12.46, P<0.01) (Figure 5). These results suggested that the cfa gene expression was increased in the tolerant strain. It is possible that the mutation of the cfa gene leads to higher stability of its expression.
Discussion
The failure of antibiotic treatment is largely attributed to drug resistance. Many mechanisms of drug resistance have been elucidated, such as the role of bacterial efflux pumps, the decrease of cell membrane permeability, the change of antibiotic targets and the modification of enzymes.14 However, another mechanism employed by bacteria to combat antibiotics is tolerance.15 Thus, it is very important to find tolerance in the clinic, delaying the occurrence of drug resistance to some extent.
To obtain a tolerant strain, we induce Shigella tolerant to ampicillin by exposing them with high-level antibiotic. Studies have reported that bacteria undergo two mutations from susceptibility to resistance: one is tolerance mutation, and the second mutation is resistance mutation on the basis of tolerance mutation, and bacterial tolerance is prior to the occurrence of drug resistance.16 Preventing the evolution of tolerance may offer a new strategy for delaying the emergence of resistance. Tolerance was assessed by measuring MDK, for example, the minimum duration of treatment that is required to kill 99% of bacteria (MDK99). A high tolerance translates to a longer MDK99, that is, a treatment of longer duration is needed to reach the same level of killing. Mechler et al17 showed that S. aureus was exposed to a lethal dose (100 MIC) of daptomycin. After 6 cycles, the drug could not effectively kill the bacteria, and the tolerant strain D6 was obtained. The MDK99 was ≥96 hrs. In our study, mel-ss2014011 (wild-type strain) was exposed to a lethal dose of ampicillin (10 MIC). After 20 cycles, bacteria could not be killed by ampicillin effectively and were tolerant to ampicillin, and we named this strain as mel-ss2014011/20. The MIC of two Shigella strains remained unchanged. The MDK99 of wild-type strain was 10 mins (from 6.5×1010 CFU/mL to 2.9×108 CFU/mL); the MDK99 of induced strain was ≥10 hrs (from 2.8×109 CFU/mL to 5×107 CFU/mL). These results suggest that the effectiveness of antibiotics was reduced and the time to treat tolerant bacteria was prolonged.
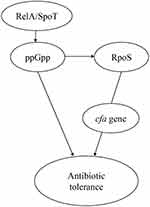
The mechanism of bacterial tolerance to antibiotics remains largely unknown. To discover the genetic basis of tolerance, wild-type strain was subjected to whole-genome sequencing and the tolerant strain was sequenced to screen non-SNPs and Indels. Compared with mel-ss2014011, mel-ss2014011-20 had one nonsynonymous mutation at cfa gene, encoding cyclopropane-fatty-acyl-phospholipid synthase, and 10 Indels were located in the noncoding region. Cyclopropane fatty acids (CFAs) are an important component of phospholipids in many bacteria15,18 and are mainly produced at the end of the exponential growth phase and right before the stable growth phase.19 Bacteria have a complex regulatory network that recognizes and regulates many physiological stresses. In Gram-negative bacteria, this regulation is through the RpoS. The RpoS is a sigma factor that can positively regulate many genes in a stationary phase,20 one of which is the cfa gene21 (Figure 6), which encodes a cyclopropane-fatty-acyl-phospholipid synthase. Murakami et al8 showed that the rpoS-knockout strain and the wild-type strain were exposed to biapenem, and the survival rate of the rpoS-knockout strain was lower than that of the wild-type strain in the stationary-growth stage, proving that the rpoS is involved in antibiotic tolerance (Figure 6), but the specific mechanism is still unclear. RpoS mainly regulates transcription, translation and post-translational levels through regulatory factors such as ppGpp, H-NS and clpP22,23 (Figure 6). Dalebroux et al showed that ppGpp is generated by relA and SpoT24 (Figure 6). Published studies have shown that the rpoS and ppGpp are interrelated, and when bacteria are exposed to antibiotics, they can keep the bacteria alive.25 The increase in RpoS mRNA translation is through ppGpp.26 In addition, the hipA mutation (G22S, D291A) can greatly increase the tolerance level in E. coli. It was found that hipA mutation could lead to the synthesis of ppGpp. The increase of ppGpp level can significantly increase the level of bacterial tolerance27 (Figure 6). In our study, the results of comparative genomics showed that the tolerant Shigella strain had one nonsynonymous mutation at cfa (G to A/Val to Met) gene, and 10 Indels were located in the noncoding region of different genes. Since the Indels are located in the noncoding region and none of these genes carry any resistance gene, these Indels are less likely to affect the tolerance of ampicillin to Shigella. It is possible that the cfa gene under the regulation of rpoS may be associated with tolerance, and the mutation of cfa gene may increase ppGpp, thereby increasing the tolerance level of Shigella to ampicillin.
We used RT-PCR to detect the expression of cfa gene. Studies have shown that cfa increases in bacteria when it responds to low pH values, as well as high temperatures, and osmotic imbalance.28 Shabala et al showed that in cfa-deletion mutations, E. coli increased proton permeability and reduced the ability to excrete H+ and they confirmed that cfa can protect E. coli from acid stress by reducing cell membrane permeability to H+.29 Macdonald et al have confirmed that an increase in cfa leads to an increase in cell membrane stability and rigidity, and an increase in cell membrane rigidity is associated with a decrease in proton permeability.30 Studies have shown that the decrease of cell membrane permeability of bacteria will lead to the decrease the concentration of antibiotics entering biofilms , the slow growth of bacteria, which will lead to bacterial tolerance to antibiotics.31–33 In our study, by measuring the tolerance level of the mutant strain, the MDK99 was 60-fold higher than the wide-type strain, indicating that the tolerance level has been reached. Real-time PCR results showed that the relative expression levels of cfa were 7.56-fold higher in the tolerant strain than in the wild-type strain. Based on the above studies, we propose a hypothesis that tolerance may be related to cfa gene expression. It is possible that the mutation of the cfa gene leads to higher stability of its expression. Increased relative expression of cfa gene in the tolerant strain may increase the rigidity of the cell membrane and decrease the permeability of the cell membrane, resulting in the inability of ampicillin to effectively kill Shigella bacteria and prolong the treatment time to make the bacteria tolerant.
cfa gene may be related to tolerance, and the mutation of the cfa gene leads to higher stability of its expression. Moreover, the cfa gene may be a target for overcoming drug tolerance, delaying the occurrence of drug resistance to some extent, and provide novel strategies for the prevention of drug resistance of Shigella.
Acknowledgments
The work was funded by the National Science and Technology Specific Projects (2018ZX10301407), Key Scientific Research Projects in Colleges and Universities of Henan Province (18B330002), Startup Research Fund of Zhengzhou University (32210273), Henan Province University Science and Technology Innovation Talent Projects (17HASTIT045).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Penatti MPA, Hollanda LM, Nakazato G. Epidemiological characterization of resistance and PCR typing of Shigella flexneri and Shigella sonnei strains isolated from bacillary dysentery cases in Southeast Brazil. Braz J Med Biol Res. 2007;40(2):249–258. doi:10.1590/S0100-879X2007000200012
2. Wang XY, Tao FB, Xiao DL. Trend and disease burden of bacillary dysentery in China (1991–2000). B World Health Organ. 2006;84(7):561–568. doi:10.2471/BLT.05.023853
3. Chukwujekwu JC, Van Heerden FR, Van Staden J. Synergistic properties of sesquiterpene lactones isolated from Centratherum punctatum Cass. in combination with ampicillin against beta-lactam-resistant Gram-negative bacteria. S Afr J Bot. 2018;117:79–82. doi:10.1016/j.sajb.2018.04.003
4. Conlon BP, Nakayasu ES, Fleck LE, et al. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503(7476):365. doi:10.1038/nature12790
5. Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi:10.1038/nrmicro1557
6. Wolfson JS, Hooper DC, McHugh GL, Bozza MA, Swartz MN. Mutants of Escherichia coli K-12 exhibiting reduced killing by both quinolone and 3-Lactam antimicrobial agents. Antimicrob Agents Chemother. 1990;34(10):1938–1943. doi:10.1128/aac.34.10.1938
7. Brauner A, Fridman O, Gefen O, Balaban NQ. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nat Rev Microbiol. 2016;14(5):320–330. doi:10.1038/nrmicro.2016.34
8. Murakami K, Ono T, Viducic D, et al. Role for rpoS gene of Pseudomonas aeruginosa in antibiotic tolerance. FEMS Microbiol Lett. 2005;242(1):161–167. doi:10.1016/j.femsle.2004.11.005
9. Kaspy I, Rotem E, Weiss N, Ronin I, Balaban NQ, Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nat Commun. 2013;4:3001. doi:10.1038/ncomms4001
10. Aaron SD, Ferris W, Ramotar K, Vandemheen K, Chan F, Saginur R. Single and combination antibiotic susceptibilities of planktonic, adherent, and biofilm-grown Pseudomonas aeruginosa isolates cultured from sputa of adults with cystic fibrosis. J Clin Microbiol. 2002;40(11):4172–4179. doi:10.1128/jcm.40.11.4172-4179.2002
11. Clinical and Laboratoy Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
12. Fridman O, Goldberg A, Ronin I, Shoresh N, Balaban NQ. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513(7518):418–421. doi:10.1038/nature13469
13. BLAST Database [homepage on the Internet]. National Institutes of Health. Available from: http://blast.ncbi.nlm.nih.gov/Blast.cgi.
14. Christopher W. Molecular mechanisms that confer antibacterial drug resistance. Nature. 2000;406(6797):775–781.
15. Wang AY, Cronan JE. The growth phase-dependent synthesis of cyclopropane fatty acids in Escherichia coli is the result of an RpoS(KatF)-dependent promoter plus enzyme instability. Mol Microbiol. 1994;11(6):1009–1017. doi:10.1111/j.1365-2958.1994.tb00379.x
16. Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N, Balaban NQ. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355(6327):826–830. doi:10.1126/science.aaj2191
17. Mechler L, Herbig A, Paprotka K, Fraunholz M, Nieselt K, Bertram R. A novel point mutation promotes growth phase-dependent daptomycin tolerance in Staphylococcus aureus. Antimicrob agents chemother. 2015;59(9):5366–5376. doi:10.1128/AAC.00643-15
18. Grogan DW, Cronan JE. Cyclopropane ring formation in membrane lipids of bacteria. Microbiol Mol Biol R. 1997;61(4):429–441.
19. Guillot A, Obis D, Mistou MY. Fatty acid membrane composition and activation of glycine-betaine transport in Lactococcus lactis subjected to osmotic stress. Int J Food Microbiol. 2000;55(1):47–51.
20. Loewen PC, Hengge-Aronis R. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu Rev Microbiol. 1994;48:53–80. doi:10.1146/annurev.mi.48.100194.000413
21. Charoenwong D, Andrews S, Mackey B. Role of rpoS in the development of cell envelope resilience and pressure resistance in stationary-phase Escherichia coli. Appl Environ Microb. 2011;77(15):5220–5229. doi:10.1128/AEM.00648-11
22. Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. doi:10.1146/annurev.micro.54.1.499
23. Van DC, Comte R, Bally AM. Stringent response activates quorum sensing and modulates cell density-dependent gene expression in Pseudomonas aeruginosa. J Bacteriol. 2001;183(18):5376–5384.
24. Dalebroux ZD, Swanson MS. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol. 2012;10(3):203–212. doi:10.1038/nrmicro2720
25. Liu S, Wu N, Zhang S, Yuan Y, Zhang W, Zhang Y. Variable persister gene interactions with (p)ppGpp for persister formation in Escherichia coli. Front Microbiol. 2017;8:1795. doi:10.3389/fmicb.2017.01795
26. Brown L, Gentry D, Elliott T, Cashel M. DksA affects ppGpp induction of RpoS at a translational level. J Bacteriol. 2002;184(16):4455–4465. doi:10.1128/jb.184.16.4455-4465.2002
27. Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2018;172(5):1135. doi:10.1016/j.cell.2018.02.023
28. Montanari C, Sado KSL, Serrazanetti DI, Etoa FX, Guerzoni ME. Synthesis of cyclopropane fatty acids in Lactobacillus helveticus and Lactobacillus sanfranciscensis and their cellular fatty acids changes following short term acid and cold stresses. Food Microbiol. 2010;27(4):493–502. doi:10.1016/j.fm.2009.12.003
29. Shabala L, Ross T. Cyclopropane fatty acids improve Escherichia coli survival in acidified minimal media by reducing membrane permeability to H+ and enhanced ability to extrude H+. Res Microbiol. 2008;159(6):458–461. doi:10.1016/j.resmic.2008.04.011
30. Macdonald PM, McDonough B, Sykes BD, McElhaney RN. Fluorine-19 nuclear magnetic resonance studies of lipid fatty acyl chain order and dynamics in Acholeplasma laidlawii B membranes. Effects of methyl-branch substitution and of trans unsaturation upon membrane acyl-chain orientational order. Biochemistry. 1983;22(22):5103–5111. doi:10.1021/bi00291a009
31. Acker HV, Dijck PV, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends Microbiol. 2014;22(6):326–333. doi:10.1016/j.tim.2014.02.001
32. Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol. 2015;34(5):877–886. doi:10.1007/s10096-015-2323-z
33. Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41(3):276–301. doi:10.1093/femsre/fux010
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.