Back to Journals » International Journal of General Medicine » Volume 14
The RNA N6-Methyladenosine Demethylase FTO Promotes Head and Neck Squamous Cell Carcinoma Proliferation and Migration by Increasing CTNNB1
Authors Zhang Y, Chen L , Wu X, Sun Z, Wang F, Wang B, Dong P
Received 15 September 2021
Accepted for publication 15 November 2021
Published 24 November 2021 Volume 2021:14 Pages 8785—8795
DOI https://doi.org/10.2147/IJGM.S339095
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Yu Zhang,1 Lixiao Chen,1 Xiaoliang Wu,2 Zhenfeng Sun,1 Fei Wang,1 Baoxin Wang,1 Pin Dong1
1Department of Otolaryngology, Head and Neck Surgery, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China; 2Tailai Bioscience, Shenzhen, People’s Republic of China
Correspondence: Pin Dong
Department of Otolaryngology, Head and Neck Surgery, Shanghai General Hospital, Shanghai Jiao Tong University School of Medicine, No. 85 Wujin Road, Hongkou District, Shanghai, 200080, People’s Republic of China
Email [email protected]
Objective: In this study, we aimed to investigate the role of RNA N6-methyladenosine demethylase fat mass and obesity-associated protein (FTO) in head and neck squamous cell carcinoma (HNSCC).
Methods: Clinical data downloaded from The Cancer Genome Atlas (TCGA) database were used to analyze the relationship between mRNA levels of FTO, METTL3, METTL14, and ALKBH5, and the overall survival in cancer and para-cancer datasets. FTO expression in tumor and normal tissues was compared using immunohistochemistry, and its relationship with overall survival was analyzed based on the Kaplan–Meier method. The FaDu cell line with high FTO levels was chosen from five HNSCC cell lines for further experiments. FTO was verified as an oncogene in HNSCC by in vitro loss-of-function and overexpression studies, cell proliferation assay, wound healing assay, and identification of expression changes of epithelial–mesenchymal transition (EMT)-related markers. Catenin beta 1 (CTNNB1) was confirmed as a downstream target gene of FTO with additional methods like the GEPIA online tool, qRT-PCR, Western blotting, and dot blot assay.
Results: We found that FTO expression was significantly upregulated in HNSCC datasets and tissues. Increased FTO expression indicated a trend towards poor prognosis and was found to promote disease proliferation and migration. Mechanistically, cell proliferation assay, wound healing assay, and identification of expression changes of EMT-related markers demonstrated that FTO could act as an oncogene in HNSCC. FTO expression was significantly correlated with CTNNB1 expression. Moreover, it exerted a tumorigenic effect by increasing CTNNB1 expression in an m6A-dependent manner.
Conclusion: FTO promotes head and neck squamous cell carcinoma proliferation and migration by increasing CTNNB1 in an m6A-dependent manner.
Keywords: FTO, m6A, HNSCC, CTNNB1
Introduction
Head and neck squamous cell carcinoma (HNSCC) is the sixth most common type of malignant tumor in the world, with approximately 600,000 newly diagnosed cases per year. The reason for its high mortality is HNSCC recurrence and metastasis, and most patients are diagnosed with HNSCC at an advanced stage.1,2 Therefore, exploring the molecular mechanisms regulating HNSCC recurrence and metastasis is likely to be of great significance for improving patient prognosis.
N6-methyladenosine (m6A) modification, which is methylation at the nitrogen-6 position of adenosine bases in mRNA, is reversible and occurs widely (>25%) as a post-transcriptional modification of mRNA in eukaryotes.3 This modification has an important role in regulating RNA transactions, including RNA splicing, stability, transport, and translation.4–7 The m6A modification is mainly mediated by the “writers” methyltransferase-like (METTL3/14),8–10 Wilms tumor 1-associated protein (WTAP),11 RNA binding motif protein 15/15B (RBM15/15B),12 and KIAA1429,13 whereas “erasers” include fat mass and obesity-associated (FTO)14 and alkB homolog 5 (ALKBH5).15 “Readers” that recognize m6A methylated transcripts include YTH domain family proteins (YTHDF1-3, YTHDC1-2), heterogeneous nuclear ribonucleoprotein (HNRNP), and IGF2 mRNA binding proteins (IGF2BPs).16,17 The m6A modification was proven to regulate the occurrence and development of various cancers and was found to play an important role in stem cell differentiation and embryonic development.4,5 To date, some studies have showed that the alterations of m6A on specific RNA sites in HNSCC and several m6A regulatory genes could influence the tumorigenesis and act as prognostic biomarkers.18,19 Positive correlation was found between selected RNA methylation machinery gene expression and m6A abundance on total RNA.20 RBM15, as a m6A-related regulator, was significantly upregulated in Laryngeal squamous cell carcinoma and relevant to the prognosis.21 Identified as m6A regulatory gene family, the expression of IGF2BP family could be an important role in tumor growth of HNSCC.22 However, relevant mechanisms of m6A RNA modification in HNSCC remain largely unknown.
FTO was first identified as a gene associated with obesity and energy metabolism and recently was proven to be the RNA m6A eraser.23 The specific role of FTO in tumor recurrence and metastasis varies among different types of cancers. Some studies have shown that FTO has tumor suppressor activity and is inversely correlated with tumor development in renal cell carcinoma, intrahepatic cholangiocarcinoma, hepatocellular carcinoma, ovarian cancer, and colorectal cancer.24–28 In contrast, FTO has also been proven to play an oncogenic role in acute leukemia, cervical cancer, melanoma, and breast cancer through its mRNA maturation regulatory function.4,29–31 However, the role of FTO in HNSCC as an m6A demethylase remains poorly studied.
In this study, we first demonstrated that FTO expression was upregulated in HNSCC tissues and that increased FTO expression indicated a trend towards poor prognosis. Then, we ascertained that FTO promotes tumor proliferation and migration in vitro by increasing the expression of the downstream target gene CTNNB1 by increasing its mRNA stability. Overall, our findings indicate the critical role of FTO in HNSCC proliferation and migration.
Materials and Methods
Bioinformatics Analysis
Clinical data, comprising that of 44 normal and 520 HNSCC tissues, were downloaded from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). The data were used to analyze the relationship between FTO, METTL3, METTL14, and ALKBH5 mRNA levels; the overall survival in cancer and para-cancer datasets was used by the UALCAN database (http://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl). We used the GEPIA website (http://gepia.cancer-pku.cn/) for correlation analysis between FTO and CTNNB1 in HNSCC.
Clinical Samples
Fifty-eight samples of hypopharyngeal and laryngeal squamous cell carcinomas, and adjacent normal tissues were collected from January 2016 to June 2019 at the Shanghai General Hospital. The samples were frozen at −80°C before RNA extraction and immunohistochemistry (IHC) analysis. We compared the expression levels of FTO in tumor tissues and normal tissues according to IHC results. The relationship between overall survival and the expression level of FTO was also analyzed. This study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of Shanghai General Hospital (2016KY175-4). Patients had given their signed informed consent prior to the study.
Immunohistochemistry Assay
The tissue was first embedded in paraffin. Next, a paraffin microtome was used to cut the tissue to sections with uniform thickness, and the sections were fixed on a clean glass slide. The sections were put into xylene and graded concentrations of alcohol for the removal of paraffin. After dewaxing was completed, sections were cleaned and treated in citrate buffer for antigen retrieval. Next, 3% H2O2 was added for 10 min at 25°C to eliminate the endogenous peroxidase activity. The sections were rinsed with distilled water and PBS, and then were blocked with 5–10% normal goat serum (diluted in PBS). Subsequently, they were incubated with the primary antibody at 4°C overnight. Following washing in PBS, sections were incubated with secondary antibody at 25°C for 20 min. After an additional wash with PBS, sections were treated with an appropriate amount of alkaline phosphatase-labeled streptavidin working solution at 37°C for 10–30 min. The color was developed with the developer for 3–15 min and the sections were rinsed with distilled water, followed by counterstaining, dehydration, transparentizing, and mounting. Evaluation of FTO expression was performed with the histological scoring method by three independent pathologists using the light microscope. To evaluate the IHC staining, the staining intensity was stratified into four levels, namely, 0, 1, 2, and 3, which were denominated as absent, weak, moderate, and strong signals, respectively. The percentage of stained cells was recorded (0% to 100%). The final IHC score was the sum of each staining intensity multiplied by the corresponding percentage, and the score scale is 0 to 300. The classification of high and low FTO expression was carried out according to the median score (≤150).
Cell Culture and Transfection
Head and neck squamous cell carcinoma cell lines, Cal27, FaDu, Tu686, HN6, and Hep-2, were purchased from the Cell Bank of the Type Culture Collection of the Chinese Academy of Science, Shanghai Institute of Cell Biology. The cell lines were maintained in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA) and 100 U/mL penicillin-streptomycin (Gibco) in a humidified atmosphere with 5% CO2 at 37°C. Lentivirus for shFTO-1 (TRCN0000246247), shFTO-2 (TRCN0000246248), FTO overexpression plasmid, double mutant (R316Q/R322Q) FTO plasmid as well as their controls were packaged with pMD2.G and psPAX2 (purchased from Addgene, Watertown, MA, USA). Briefly, 1.5 μg pMD2.G, 3 μg psPAX2, and 4.5 μg construct for overexpression or knockdown of specific genes were co-transfected into HEK-293T cells in 100 mm cell culture dish with Effectene Transfection Reagent (301427, QIAGEN, Valencia, CA). The lentivirus particles were harvested at 48 and 72 hours after transfection. Finally, the lentivirus particles were directly added into target cells before incubating at 37°C for 24–48 hours. Lastly, puromycin (1 ug/mL) was added to the cells for 5 days for establishing stable cell lines.
RNA Extraction, Reverse Transcription, and Amplification
Total RNA was extracted from tissues and cell lines using RNAiso Plus (code No. 9108, Takara, Shiga, Japan). The RT-PCR kit (code No. RR036A, Takara) was used to reverse transcribe total RNA into cDNA according to the manufacturer’s protocol. Quantitative real-time fluorescence quantitative polymerase chain reaction (qRT-PCR) was performed using the TB Green PCR Premix Ex TaqTM II Kit (code no. RR820A, Takara) on an Applied Biosystems 7500 device (Foster City, CA, USA). All qRT-PCR tests were repeated three times. The standard for gene expression was determined based on the expression of the internal control GAPDH. The primer sequences were as follows: GAPDH forward primer, 5ˈ-ACAACTTTGGTATCGTGGAAGG-3ˈ; reverse primer, 5ˈ-GCCATCACGCCACAGTTTC-3ˈ; METTL3 forward primer, 5ˈ-TTGTCTCCAACCTTCCGTAGT-3ˈ; reverse primer, 5ˈ-CCAGATCAGAGAGGTGGTGTAG-3ˈ; METTL14 forward primer, 5ˈ-AGTGCCGACAGCATTGGTG-3′; reverse primer, 5ˈ-GGAGCAGAGGTATCATAGGAAGC-3ˈ; FTO forward primer, 5ˈ-GCTGCTTATTTCGGGACCTG-3ˈ; reverse primer, 5ˈ-AGCCTGGATTACCAATGAGGA-3ˈ; ALKBH5 forward primer, 5ˈ-CGGCGAAGGCTACACTTACG-3ˈ; reverse primer, 5ˈ-CCACCAGCTTTTGGATCACCA-3ˈ; CTNNB1 forward primer, 5ˈ-CTGAGGAGCAGCTTCAGTCC-3ˈ; reverse primer, 5ˈ-GGCCATGTCCAACTCCATCA-3ˈ; m6A PCR forward primer, 5ˈ-CAGGGA-GAACCCCTTGGATA-3ˈ; reverse primer, 5ˈ-GTTCCATCATGGGGTCCATA-3ˈ.
Western Blotting
RIPA lysis buffer (PC101, EpiZyme, Shanghai, China) was used to extract the proteins from cells. Protein concentrations were determined using a bicinchoninic acid assay reagent kit (Beyotime, Shanghai, China). Protein lysates were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and subsequently transferred onto a polyvinylidene fluoride (PVDF, WJ002, EpiZyme) membrane. The PVDF membrane was blocked with 5% skimmed milk at 25°C for 2 h followed by overnight incubation with primary antibodies (anti-FTO, 1:1000, Abcam, USA, ab92821; anti-GAPDH, 1:1000, Abcam, USA, ab8245; anti-CTNNB1 1:1000;8480S; Cell Signaling Technology, USA, anti-slug 1:2000, Abcam, USA, ab51772; anti-ZEB1, 1:500, Abcam, USA, ab203829; anti-SOX2, 1:500, Abcam, USA, ab92494) at 4°C. The membrane was then incubated with secondary antibodies at 25°C for 2 h and binding signals were detected using an enhanced chemiluminescent kit (Beyotime, China). Afterwards, a multi-imaging system (Tanon 5200, Tanon Science & Technology Inc., Shanghai, China) was used for image acquisition and ImageJ program was used for data quantification.
Measurement of m6A Levels
Total RNA from tissues and cell lines was extracted using RNAiso Plus (code no. 9108, Takara, Shiga, Japan) with deoxyribonuclease I (Sigma, Shanghai, China). The m6A RNA methylation quantification kit (ab185912; Abcam, USA) was used to detect m6A levels in the total RNA as per the manufacturer’s protocol. The m6A level was determined by measuring the absorbance at 450 nm (Thermo Fisher, USA).
Cell Proliferation Assay
Cells were added to a 96-well plate at approximately 1500 cells per well, followed by incubation in a humidified atmosphere of 5% CO2 at 37°C. A cell counting kit-8 (CCK-8; Obio Technology, Shanghai, China) was used for cell proliferation assays as per the manufacturer’s protocol, and the absorbances at 450 nm were noted at 0, 24, 48, 72, and 96 h.
Wound Healing Assay
The digested cells were plated with appropriate confluence to ensure that the confluence was close to 100% overnight. A linear wound was scratched along the surface by a 10ul pipette tip. The serum-free medium was added for incubation in a humidified atmosphere of 5% CO2 at 37°C after the cells were rinsed with PBS. At 0 and 24 hours after the incubation, the images were acquired with an inverted microscope after the incubation and analyzed with image J. The percentage of wound closure (%) = (width on day 0 - with on day 24)/width on day 0 × 100%.
Dot Blot Assay
The procedures were carried out according to the operation manual. The cell clone solution was fixed on the nitrocellulose membrane, and was dried and incubated with sealing solution at 25°C for 1 h. The nitrocellulose membrane was soaked in the primary antibody and incubated at room temperature for 1 h, and the nitrocellulose film was then soaked in the second antibody again after washing with TBST solution. It was then incubated at room temperature for 1 h and then washed in TBST solution again. Finally, the nitrocellulose film was immersed in fluorescent chromogenic reagent and exposed.
Statistical Analysis
All experiments were repeated three times and GraphPad Prism (version 8; GraphPad Software, Inc., San Diego, CA, USA) was used for statistical analysis. Data are presented as the mean ± SD. The significance of differences between two groups was determined using a Student’s t-test. One-way ANOVA was performed to compare the results of more than two groups. Data were considered statistically significant at P < 0. 05.
Results
Prognostic Value of m6A Methyltransferases and Demethylases in HNSCC
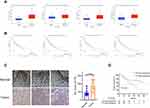
m6A modification is a dynamic process mediated by genes known as writers and erasers.32,33 To investigate the potential roles of m6A modulators, which might result in poor clinical outcomes in HNSCC, we first compared m6A modulator transcriptome expression between normal and tumor tissues in TCGA. We found that all four m6A modulators showed higher mRNA expression in HNSCC tissues (Figure 1A). Furthermore, we analyzed their prognostic values in TCGA cohort. Based on the Kaplan–Meier method and the Log rank test, we found that patients with high FTO expression had a trend towards poor prognosis. Although there was no significant difference in this correlation, with P > 0.05, the relationship between FTO and prognosis was relatively more important than that of the other three genes (Figure 1B).
We also performed an IHC staining assay to investigate FTO protein expression levels in tumor and adjacent normal tissue pairs from patients with HNSCC (Figure 1C). Consistent with the results based on the TCGA database, FTO was expressed significantly higher in HNSCC tissues compared to normal tissues, according to IHC results (Figure 1C). We further conducted survival analysis based on the expression level of FTO and the overall survival of patients; the results showed that the overall survival of patients with high FTO expression was significantly lower than that of patients with low FTO expression (Figure 1D). In addition, we found from Table 1 that there exists a correlation between smoking status and FTO expression. Among the 37 patients with a non-smoking history, the proportion of patients with low FTO expression was higher. Meanwhile, the proportion of patients with high FTO expression was higher in 21 patients with a history of smoking. The relevant clinical information of patients is shown in Table 1.
 |
Table 1 Summary of Patient Characteristics |
FTO Acts as an Oncogene in HNSCC
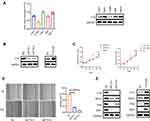
To investigate the relationship between FTO and the phenotype of HNSCC, we first evaluated the mRNA and protein levels of FTO in five HNSCC cell lines, Cal27, FaDu, Tu686, HN6 and Hep-2 (Figure 2A), and chose the FaDu cell line with high FTO levels for further experiments. Based on loss-of-function studies in vitro, we knocked down FTO and constructed stable FTO-overexpressing FaDu cell lines (Figure 2B). The proliferation rate of FaDu cells was effectively retarded by FTO knock-down and accelerated by FTO overexpression (Figure 2C). Based on wound healing assay, FTO knockdown inhibited cell migration; however, FTO overexpression enhanced cell migration in FaDu cell line (Figure 2D). In addition, FTO knockdown decreased the expression of epithelial–mesenchymal transition (EMT)-related markers and FTO overexpression increased EMT-related markers protein levels (Sox2, Slug, and ZEB1; Figure 2E).
CTNNB1 is a Downstream Target Gene of FTO
To identify the underlying mechanisms by which FTO is involved in HNSCC progression and metastasis, a previous study reported that FTO could demethylate CTNNB1 transcripts and promote CTNNB1 expression in cervical squamous cell carcinoma.34 First, as we expected, FTO expression was significantly correlated with CTNNB1 expression in TCGA database (Figure 3A). FTO knockdown increased m6A levels, and replenishing wild-type (WT) FTO but not mutant FTO decreased m6A levels in the FaDu cell line, based on a dot blot assay (Figure 3B). By qRT-PCR, we found that CTNNB1 was remarkably inhibited by FTO deficiency in FaDu cells (Figure 3C). CTNNB1 mutations, via constitutive activation of the Wnt signaling pathway, are involved in the development of cancer. Next, we constructed WT FTO- and mutant FTO-overexpression FaDu cells and found that WT FTO significantly increased CTNNB1 expression, but not the mutant FTO (Figure 3D). By performing gene-specific m6A qRT-PCR validation, after FTO knockdown in HNSCC cells, m6A levels were significantly increased compared with those in normal control cells (Figure 3E).
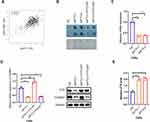 |
Figure 3 (A) The correlation between FTO and CTNNB1 RNA expression levels in the TCGA dataset for HNSCC assessed by using the GEPIA online tool (http://gepia.cancer-pku.cn/). (B) m6A dot blot assay of knockdown and replenishment of wild-type (WT) and mutant (MT) FTO in FaDu cells. Methylene blue staining was used as the control. (C) RT-qPCR arrays were used to detect the RNA expression levels of CTNNB1 after FTO knockdown in FaDu cell, normalized to GAPDH. Data are presented as the mean ± SD; **P < 0.01. (D) CTNNB1 RNA expression levels were analyzed in FTO-WT and FTO-mutant FaDu cells, normalized to GAPDH. Immunoblot assay of CTNNB1 protein levels in FaDu cells upon FTO knockdown and FTO-WT and FTO-mutant. Data are presented as the mean ± SD, **P < 0.01. (E) m6A RIP and qRT-PCR analysis of m6A levels in CTNNB1 mRNA in shFTO-1/2 or corresponding wild-type cells. Data are expressed as the mean ± SD; *P < 0.05. Abbreviations: ns, not significant; FTO, fat mass- and obesity-associated protein; CTNNB1, Catenin beta 1. |
CTNNB1 Reverses the Effects of FTO in HNSCC Cell Lines
We then explored whether CTNNB1 could reverse the effects of FTO deficiency and overexpression on HNSCC cell proliferation and migration. We first constructed CTNNB1-knock-down FaDu cell line and verified that CTNNB1 deficiency could inhibit FaDu cell growth and migration (Figure 4A and B). Then, we overexpressed CTNNB1 in the FTO-knockdown FaDu cell line. As expected, CTNNB1 overexpression notably reversed the inhibition of HNSCC proliferation and migration resulting from FTO deficiency (Figure 4C and D). Together, these results showed that that CTNNB1 is the downstream target gene of FTO and that FTO could accelerate HNSCC proliferation and migration by increasing CTNNB1 expression.
Discussion
N6-methyladenosine RNA modification is a post-transcriptional regulation of gene expression, which has wide-ranging effects on biological processes.35–38 The m6A modification has been shown to be involved in the occurrence and progression of various cancers by regulating different downstream genes.16,39,40 Several studies till date have focused on the role of m6A regulatory genes in HNSCC,18–22 especially on prognosis prediction and guidance for therapeutic strategies selection and yet, little is known about the underlying mechanisms of m6A RNA modification in HNSCC. As RNA m6A eraser, FTO is involved in the demethylation of m6A in various cancers. For example, in ovarian cancer, FTO plays a tumor suppressor role by constraining cancer stem cell self-renewal.24 In hepatocellular carcinoma and intrahepatic cholangitis carcinoma, FTO was proven to inhibit tumorigenesis.25,26 However, FTO promotes glioblastoma tumorigenesis and cancer stem cell renewal by regulating target genes.41 FTO, via the m6A regulatory machinery, degrades the pro-apoptosis gene BNIP3, resulting in the promotion of breast cancer progression.42 In addition to these conflicting studies about the different roles in various cancers, the function of FTO in HNSCC was unknown to date. Our report showed that FTO exerts an oncogenic effect on HNSCC progression, meaning that FTO might be an important marker for predicting HNSCC progression. Combined with our previous work on NSUN2,43 these findings lead us to propose a mechanism through which mRNA modification affects the tumorigenesis, development, and metastasis of HNSCC. The β-catenin protein encoded by the CTNNB1 gene is an important effector of the Wnt signaling pathway, and the pathway can regulate cell cycle, cell apoptosis, cell transformation and other processes, participating in tumorigenesis and development.44 CTNNB1 ectopic expression exists in liver cancer, gastric cancer, colon cancer, and breast cancer with high incidence.45 CTNNB1, combining with T cell factor/lymph enhancer factor, can enter the cell nucleus and activate genes that promote cancer proliferation, thereby increasing the malignant behavior of cervical cancer cells.46 Kurnit et al show that CTNNB1 gene mutations can predict the risk of endometrial cancer recurrence, suggesting that CTNNB1 may be related to tumor recurrence.47
In our study, we found that FTO was highly expressed in HNSCC compared to that in normal tissues and that patients with high FTO expression had a trend toward poor prognosis, indicating the role of FTO in the regulation of HNSCC development. Regarding the relationship between smoking status and FTO expression, we found that among patients with a history of smoking, there were more patients with high FTO expression. Smoking has been considered an unfavorable prognostic factor for HNSCC.48–50 Therefore, high FTO expression in patients with a history of smoking was compatible with our research view that patients with high FTO expression had a trend towards poor prognosis. A recent study on lung cancer mentioned that some m6A regulatory factors, including FTO, were related to smoking status.51 In later studies, we will continue to focus on the potential relationship between smoking status and FTO in HNSCC. We also designed serial biological functional assays using the FaDu cell line to demonstrate that FTO plays a pivotal role in promoting cell proliferation and metastasis in vitro. To explore the molecular mechanism by which FTO promotes tumor growth in HNSCC, we analyzed TCGA database for HNSCC. We found that the expression level of FTO was significantly correlated with one oncogenic transcription factor, CTNNB1. We then performed qRT-PCR and Western blot experiments, whose results suggest that FTO knockdown in the FaDu cell line would lead to a decrease in the mRNA and protein level of CTNNB1. We also investigated wild-type FTO and a control catalytically inactive mutant using an FTO-knockdown FaDu cell line, as control. We found that WT FTO, but not the inactive mutant, could stabilize mRNA and protein levels. These results indicated that FTO stabilizes the mRNA level in FaDu cells to promote tumorigenesis and the development and metastasis of HNSCC.
There are some limitations in this study, such as the lack of in vivo experiments on FTO and CTNNB1, and the lack of m6A methylation sequencing to clarify CTNNB1 as the target gene of FTO. In summary, we provide here the first report that FTO, the key m6A demethylase, is upregulated in HNSCC patients and that patients with high expression had a trend towards poor prognosis. FTO was found to promote HNSCC cell line proliferation and migration by stabilizing mRNA encoding CTNNB1, which has oncogenic roles in various cancers. Altogether, our findings suggest that FTO might serve as a novel potential therapeutic target for HNSCC.
Conclusion
Our studies demonstrated a novel mechanism that FTO promotes head and neck squamous cell carcinoma proliferation and migration by increasing CTNNB1 in an m6A-dependent manner.
Abbreviations
Ctrl, control; HNSCC, head and neck squamous cell carcinoma; IHC, immunohistochemistry; m6A, N6-methyladenosine; FTO, fat mass and obesity-associated protein; CTNNB1, catenin beta 1; qRT-PCR, quantitative real-time fluorescence quantitative PCR; TBST, 0.1% Tris-HCl with Tween-20; TCGA, The Cancer Genome Atlas; METTL3, methyltransferase -like 3; METTL14, methyltransferase-like 14; ALKBH5, alkB homologue 5; EMT, epithelial–mesenchymal transition; WTAP, Wilms tumour 1-associated protein, IGF2BP, IGF2 mRNA binding proteins; m6A, N6-methyladenosine; RBM15/15B, RNA binding motif protein 15/15B, YTHDF, YT521-B homology domain-containing protein family.
Data Sharing Statement
The original data of this article can be obtained from the corresponding author on reasonable request.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
National Natural Science Foundation of China (grant number: 82072989), Emerging advanced technology joint research project (grant number: SHDC12020120).
Disclosure
Xiaoliang Wu is an employee of Tailai Bioscience, Shenzhen, China. The authors declare that they have no other conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–386.
3. Deng X, Su R, Weng H, Huang H, Li Z, Chen J. RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517.
4. Li Z, Weng H, Su R, et al. FTO plays an oncogenic role in acute myeloid leukemia as a N(6)-Methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141.
5. Zhang C, Samanta D, Lu H, et al. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m(6) A-demethylation of NANOG mRNA. Proc Natl Acad Sci U S A. 2016;113(14):E2047–2056.
6. Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399.
7. Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife. 2017;6:e311311.
8. Lin S, Choe J, Du P, Triboulet R, Gregory RI. The m(6)A Methyltransferase METTL3 promotes translation in human cancer cells. Mol Cell. 2016;62(3):335–345.
9. Li T, Hu PS, Zuo Z, et al. METTL3 facilitates tumor progression via an m(6) A-IGF2BP2-dependent mechanism in colorectal carcinoma. Mol Cancer. 2019;18(1):112.
10. Chen X, Xu M, Xu X, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19(1):106.
11. Schöller E, Weichmann F, Treiber T, et al. Interactions, localization, and phosphorylation of the m(6)A generating METTL3-METTL14-WTAP complex. RNA (New York, NY). 2018;24(4):499–512.
12. Zhai J, Li S, Sen S, et al. m(6)A RNA methylation regulators contribute to eutopic endometrium and myometrium dysfunction in adenomyosis. Front Genet. 2020;11:716.
13. Wang P, Wang Z, Zhang M, Wu Q, Shi F, Yuan S. KIAA1429 and ALKBH5 oppositely influence aortic dissection progression via regulating the maturation of Pri-miR-143-3p in an m6A-dependent manner. Front Cell Develop Biol. 2021;9:668377.
14. Tsuruta N, Tsuchihashi K, Ohmura H, et al. RNA N6-methyladenosine demethylase FTO regulates PD-L1 expression in colon cancer cells. Biochem Biophys Res Commun. 2020;530(1):235–239.
15. Kowalski-Chauvel A, Lacore MG, Arnauduc F, et al. The m6A RNA Demethylase ALKBH5 promotes radioresistance and invasion capability of glioma stem cells. Cancers. 2020;13:1.
16. He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176.
17. Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28(6):616–624.
18. Zhao X, Cui L. Development and validation of a m(6)A RNA methylation regulators-based signature for predicting the prognosis of head and neck squamous cell carcinoma. Am J Cancer Res. 2019;9(10):2156–2169.
19. Zhou X, Han J, Zhen X, et al. Analysis of genetic alteration signatures and prognostic values of m6A regulatory genes in head and neck squamous cell carcinoma. Front Oncol. 2020;10:718.
20. Romanowska K, Rawłuszko-Wieczorek AA, Marczak Ł, Kosińska A, Suchorska WM, Golusiński W. The m(6)A RNA modification quantity and mRNA expression level of RNA methylation-related genes in head and neck squamous cell carcinoma cell lines and patients. Biomolecules. 2021;11:6.
21. Wang X, Tian L, Li Y, et al. RBM15 facilitates laryngeal squamous cell carcinoma progression by regulating TMBIM6 stability through IGF2BP3 dependent. J Exp Clin Cancer Res. 2021;40(1):80.
22. Paramasivam A, George R, Priyadharsini JV. Genomic and transcriptomic alterations in m6A regulatory genes are associated with tumorigenesis and poor prognosis in head and neck squamous cell carcinoma. Am J Cancer Res. 2021;11(7):3688–3697.
23. Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887.
24. Huang H, Wang Y, Kandpal M, et al. FTO-dependent N (6)-methyladenosine modifications inhibit ovarian cancer stem cell self-renewal by blocking cAMP signaling. Cancer Res. 2020;80(16):3200–3214.
25. Liu X, Liu J, Xiao W, et al. SIRT1 regulates N(6)-Methyladenosine RNA modification in hepatocarcinogenesis by inducing RANBP2-dependent FTO SUMOylation. Hepatology. 2020;72(6):2029–2050.
26. Rong ZX, Li Z, He JJ, et al. Downregulation of Fat Mass and Obesity Associated (FTO) promotes the progression of intrahepatic cholangiocarcinoma. Front Oncol. 2019;9:369.
27. Zhuang C, Zhuang C, Luo X, et al. N6-methyladenosine demethylase FTO suppresses clear cell renal cell carcinoma through a novel FTO-PGC-1alpha signalling axis. J Cell Mol Med. 2019;23(3):2163–2173.
28. Ruan DY, Li T, Wang YN, et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene. 2021;40(33):5168–5181.
29. Yang S, Wei J, Cui YH, et al. m(6)A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat Commun. 2019;10(1):2782.
30. Zou D, Dong L, Li C, Yin Z, Rao S, Zhou Q. The m(6)A eraser FTO facilitates proliferation and migration of human cervical cancer cells. Cancer Cell Int. 2019;19:321.
31. Liu J, Ren D, Du Z, Wang H, Zhang H, Jin Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem Biophys Res Commun. 2018;502(4):456–464.
32. Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother/Biomedecine & Pharmacotherapie. 2019;112:108613.
33. Yang G, Sun Z, Zhang N. Reshaping the role of m6A modification in cancer transcriptome: a review. Cancer Cell Int. 2020;20:353.
34. Zhou S, Bai ZL, Xia D, et al. FTO regulates the chemo-radiotherapy resistance of cervical squamous cell carcinoma (CSCC) by targeting β-catenin through mRNA demethylation. Mol Carcinog. 2018;57(5):590–597.
35. Zhou Z, Lv J, Yu H, et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol Cancer. 2020;19(1):104.
36. Williams GD, Gokhale NS, Horner SM. Regulation of viral infection by the RNA modification N6-Methyladenosine. Ann Rev Virol. 2019;6(1):235–253.
37. Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505(7481):117–120.
38. Imam H, Kim GW, Siddiqui A. Epitranscriptomic(N6-methyladenosine) modification of viral RNA and virus-host interactions. Front Cell Infect Microbiol. 2020;10:584283.
39. Yue B, Song C, Yang L, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142.
40. Lan T, Li H, Zhang D, et al. KIAA1429 contributes to liver cancer progression through N6-methyladenosine-dependent post-transcriptional modification of GATA3. Mol Cancer. 2019;18(1):186.
41. Cui Q, Shi H, Ye P, et al. m(6)A RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18(11):2622–2634.
42. Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer. 2019;18(1):46.
43. Chen L, Ding J, Wang B, et al. RNA methyltransferase NSUN2 promotes hypopharyngeal squamous cell carcinoma proliferation and migration by enhancing TEAD1 expression in an m(5) C-dependent manner. Exp Cell Res. 2021;404(2):112664.
44. Tai D, Wells K, Arcaroli J, et al. Targeting the WNT signaling pathway in cancer therapeutics. The Oncologist. 2015;20(10):1189–1198.
45. Espada J, Calvo MB, Díaz-Prado S, Medina V. Wnt signalling and cancer stem cells. Clin Transl Oncol. 2009;11(7):411–427.
46. Chen M, Li L, Zheng PS. SALL4 promotes the tumorigenicity of cervical cancer cells through activation of the Wnt/β-catenin pathway via CTNNB1. Cancer Sci. 2019;110(9):2794–2805.
47. Kurnit KC, Kim GN, Fellman BM, et al. CTNNB1 (beta-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Mod Pathol. 2017;30(7):1032–1041.
48. Ghasemi F, Prokopec SD, MacNeil D, et al. Mutational analysis of head and neck squamous cell carcinoma stratified by smoking status. JCI Insight. 2019;4:1.
49. Du E, Mazul AL, Farquhar D, et al. Long-term survival in head and neck cancer: impact of site, stage, smoking, and human papillomavirus status. Laryngoscope. 2019;129(11):2506–2513.
50. de la Iglesia JV, Slebos RJC, Martin-Gomez L, et al. Effects of tobacco smoking on the tumor immune microenvironment in head and neck squamous cell carcinoma. Clin Cancer Res. 2020;26(6):1474–1485.
51. Li N, Zhan X. Identification of pathology-specific regulators of m(6)A RNA modification to optimize lung cancer management in the context of predictive, preventive, and personalized medicine. EPMA J. 2020;11(3):485–504.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.