Back to Journals » Infection and Drug Resistance » Volume 15
The Regulation of ManLAM-Related Gene Expression in Mycobacterium tuberculosis with Different Drug Resistance Profiles Following Isoniazid Treatment
Authors Yimcharoen M, Saikaew S, Wattananandkul U , Phunpae P, Intorasoot S, Kasinrerk W, Tayapiwatana C, Butr-Indr B
Received 10 November 2021
Accepted for publication 21 January 2022
Published 5 February 2022 Volume 2022:15 Pages 399—412
DOI https://doi.org/10.2147/IDR.S346869
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Manita Yimcharoen,1 Sukanya Saikaew,1 Usanee Wattananandkul,1 Ponrut Phunpae,1 Sorasak Intorasoot,1 Watchara Kasinrerk,2,3 Chatchai Tayapiwatana,2– 4 Bordin Butr-Indr1
1Division of Clinical Microbiology, Department of Medical Technology, Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, 50200, Thailand; 2Division of Clinical Immunology, Department of Medical Technology, Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, 50200, Thailand; 3Biomedical Technology Research Center, National Center for Genetic Engineering and Biotechnology, National Science and Technology Development Agency at The Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, 50200, Thailand; 4Center of Biomolecular Therapy and Diagnostic, Faculty of Associated Medical Sciences, Chiang Mai University, Chiang Mai, 50200, Thailand
Correspondence: Bordin Butr-Indr, Tel +66 53945086 ext. 15
, Fax +66 53217143
, Email [email protected]; [email protected]
Introduction: Tuberculosis (TB) caused by Mycobacterium tuberculosis (MTB) remains a global health concern because of the development of drug resistance. The adaptability of MTB in response to a variety of environmental stresses is a crucial strategy that supports their survival and evades host defense mechanisms. Stress regulates gene expression, particularly virulence genes, leading to the development of drug tolerance. Mannose-capped lipoarabinomannan (ManLAM) is a critical component of the cell wall, functions as a virulence factor and influences host defense mechanisms.
Purpose: This study focuses on the effect of isoniazid (INH) stress on the regulation of ManLAM-related genes, to improve our understanding of virulence and drug resistance development in MTB.
Materials and Methods: MTB with distinct drug resistance profiles were used for gene expression analysis. Multiplex-real time PCR assay was performed to monitor stress-related genes (hspX, tgs1, and sigE). The expression levels of ManLAM-related genes (pimB, mptA, mptC, dprE1, dprE2, and embC) were quantified by qRT-PCR. Sequence analysis of drug resistance-associated genes (inhA, katG, and rpoB) and ManLAM-related genes were performed to establish a correlation between genetic variation and gene expression.
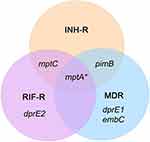
Results: INH treatment activates the stress response mechanism in MTB, resulting in a distinct gene expression pattern between drug resistance and drug-sensitive TB. In response to INH, hspX was up-regulated in RIF-R and MDR. tgs1 was strongly up-regulated in MDR, whereas sigE was dramatically up-regulated in the drug-sensitive TB. Interestingly, ManLAM-related genes were most up-regulated in drug resistance, notably MDR (pimB, mptA, dprE1, and embC), implying a role for drug resistance and adaptability of MTB via ManLAM modulation.
Conclusion: This study establishes a relationship between the antibiotic stress response mechanism and the expression of ManLAM-related genes in MTB samples with diverse drug resistance profiles. The novel gene expression pattern in this work is valuable knowledge that can be applied for TB monitoring and treatment in the future.
Keywords: Mycobacterium tuberculosis, isoniazid, antibiotic stress, ManLAM, stress response, drug resistance
Introduction
Mycobacterium tuberculosis (MTB) is a bacterium that causes tuberculosis (TB), a serious global health concern. Drug-resistant TB is a critical challenge for tuberculosis diagnosis, monitoring, and treatment. Virulence of MTB is influenced by various factors, including the natural background of the bacterium and their adaptive mechanisms in response to environmental stress.1–3 Mycobacterial cell wall serves as a barrier against the entry of harmful chemicals or antibiotic drugs into the cell.4 Mannose-capped lipoarabinomannan (ManLAM) is a glycolipid found in the cell wall of MTB that acts as a virulence factor, supports MTB survival, and prevents the host defense mechanisms.5 ManLAM influences the host immune response in several ways, including phagosome maturation, antigen presentation, T cell activation and cytokine regulation.6 The major steps in ManLAM synthesis are mannan core synthesis, arabinan domain production, and ManLAM capping,7 which require the coordination of several genes. Phosphatidylinositol mannosides (PIMs) provide the structural basis for lipomannan (LM), lipoarabinomannan (LAM), and ManLAM. pimB (Rv2188c) produce PIMs from phosphatidylinositol (PI).8 mptA (Rv2174) and mptC (Rv2181) are involved in the elongation and branching of LM, respectively.9,10 For arabinomannan domain production, dprE1 (Rv3790) and dprE2 (Rv3791) convert decaprenylphosphoryl ribose (DPR) to decaprenylphosporyl arabinose (DPA).11 embC (Rv3793) utilizes DPA as a precursor for LAM formation.12 The last step is ManLAM capping by Rv1635c13 and mptC (Rv2181) to form ManLAM.10
MTB is a successful pathogen, they utilize their adaptation strategies to survive under environmental stress, escape host-killing systems and promote TB progression. The activation of stress responder genes and virulence gene regulation in response to several types of stress are key factors contributing to drug resistance development.3,14 The DosR regulon consists of approximately 50 genes that regulate various physiological pathways, respond to several stresses, and is crucial for mycobacteria to become dormant.15 hspX (acr, Rv2031c) and tgs1 (Rv3130c) are DosR-regulon genes and play an important role as stress responder genes supporting mycobacterium survival.3,16,17 Previous research has found that the concentration of oxygen, nitric oxide, and carbon monoxide influence hspX expression.18 The combination of various stresses in the multi-stress model increases expression levels of tgs1, which encodes triacylglycerol synthase, leading to triacylglycerol accumulation and drug resistance development.3,17 sigE (Rv1221) is one of the RNA polymerase sigma factors that sense to a wide range of stress such as hypoxia,18,19 nutrient starvation20 and acidic pH.21,22 These findings suggest that hspX, tgs1, and sigE are stress-related genes that can be used to determine the stress response of mycobacteria. The emergence of drug resistance is a major obstacle for TB treatment. Inappropriate antibiotic use is a significant contributor to antibiotic resistance.1 Isoniazid (INH) is a first-line drug that has bactericidal activity and is widely used for TB treatment. INH requires the activation by redox enzymes encoded by the katG gene of MTB to be an active form. The active form binds to enoyl-acyl carrier protein reductase (InhA), resulting in inhibition of mycolic acid synthesis and cell death.23,24 Phenotypic INH-resistance is associated with mutations of several genes including, katG, inhA, ahpC, kasA, and ndh.25 Among these, katG (S315T) and inhA regulatory region (C-15T) are the most mutation points found in INH resistance.25 Previous publications26,27 provide evidence that INH may not directly target only mycolic acid but can also alter other parts of MTB. During INH treatment, MTB up-regulates several genes involved in fatty acid synthesis and degradation, the activity of peroxidase, transporters, and efflux pumps.26 INH-R and drug-sensitive clinical isolates regulate cell wall components differently following INH treatment. Phthiocerol dimycocerosate (PDIM), trehalose monomycolate (TMM), and PIMs were decreased in INH-R compared to drug-sensitive TB.28 Furthermore, INH induces the accumulation of free trehalose, which serves as a signal for the iniBAC regulator (IniR) to activate the major cell wall stress operon iniBAC.29 These findings imply that INH treatment influences the expression of genes involved in the biosynthesis of mycobacterial cell wall components. However, the adaptation ability of MTB in response to INH associated with the development of drug resistance remains unclear.
In this work, INH was utilized as the stress to induce adaptive mechanism of MTB. MTB strains including a drug-susceptible strain (H37Rv) and clinical MTB isolates, which are isoniazid resistant (INH-R), rifampin resistant (RIF-R), and multi-drug resistant (MDR), were cultured. Following the treatment of INH at CMax (6 µg/mL)30 concentration for 30 min, gene expression analysis was performed and compared among four strains. The expression of stress-related genes such as hspX, tgs1, and sigE were investigated by multiplex real-time PCR assay. The relative expression of ManLAM-related genes that are involved in ManLAM synthesis from early to late steps, including pimB, mptA, mptC, dprE1, dprE2, and embC, were determined by real-time PCR. Sequence analysis of ManLAM-related genes and drug resistance-associated genes were performed to determine the relationship between genetic background and gene expression levels. This study aimed to determine the expression of stress-related genes and ManLAM-related genes in response to INH in MTB strains with different drug resistance profiles, to improve understanding of drug resistance development in MTB.
Materials and Methods
Ethics and Biosafety Approval
This work was approved by the Institutional Ethics Committee and Biosafety Committee, Chiang Mai University (approval no.: AMSEC-63EM-028, CMUIBC A-0564001).
Mycobacterial Strains and Culture
MTB strains with diverse drug resistance profiles, including H37Rv (Drug-sensitive strain), INH-R, RIF-R, and MDR, were isolated from TB patients in northern Thailand in 2015 by the Office of Disease Prevention and Control, 1 (Chiang Mai, Thailand) and kept at −80°C. The drug susceptibility profile was tested using the BACTEC mycobacterial growth indicator tube (MGIT) 960 system as described previously.31 All mycobacteria were cultured in Lowenstein-Jensen medium (Biomedia, Thailand) at 37°C for 1 month or until further experiments were performed.
Drug Resistance Profiles
To confirm the genotypic and phenotypic drug resistance profiles of MTB samples, GenoType MTBDRplus assay (Hain Lifescience, Germany) and drug susceptibility test by agar proportion method were performed, respectively. For GenoType MTBDRplus assay, the mutations of drug resistance-associated genes such as rpoB, katG, and inhA were detected using reverse hybridization of amplicons immobilized on membranes. This assay was performed and interpreted according to the manufacturer’s guideline. The agar proportion was performed following the Clinical and Laboratory Standards Institute (CLSI) document M24-A232 using recommended critical concentrations of isoniazid, rifampin, streptomycin, and ethambutol.
Isoniazid Preparation and Treatment
Isoniazid (INH) drug was utilized as a stressor to stimulate the stress response mechanism of MTB, each mycobacterium was cultured and treated following previous publication.31 INH (Sigma-Aldrich, USA) was prepared according to a previous publication.31 A stock concentration of 1 mg/mL was prepared using sterile distilled water and sterilized by 0.2 µm nylon membrane filters, stored at −20°C prior to use. Mycobacteria were culture in 20 mL of Middlebrook 7H9 (M7H9) supplement with 0.05% (v/v) tween 20, 10% (v/v) OADC and 0.5% (v/v) glycerol at 35°C for 1 week. Prior to drug treatment, 10 mL of bacterial culture was transferred to the new tube and adjusted to McFarland standard No.1 with Middlebrook 7H9. Isoniazid at a CMax concentration (6 µg/mL)30 was added to mycobacterial culture tube, mixed and incubated at 35°C for 30 min. An untreated tube of each strain was used as control. The treated cells were pelleted by centrifugation at 12,000×g at 4°C for 20 min (Beckman Allegra X-15R) and the supernatant was discarded. The pellets were washed twice with sterile phosphate buffered solution (PBS) and centrifuged at 12,000×g at 4°C for 15 min before performing RNA extraction.
RNA Extraction and Complementary DNA (cDNA) Preparation
For gene expression analysis, RNA was extracted and converted to cDNA using Nucleospin® RNA kit (MACHEREY-NAGEL, Germany) and ReverTra Ace® qPCR RT Master Mix with gDNA remover (Toyobo, Japan), respectively. Mycobacterial RNA was extracted following the manufacturer’s protocols with some modification at the first step for cell lysis. Tris-EDTA buffer (10mM Tris-HCl, 1mM EDTA; pH 8) containing 2 mg/mL of lysozyme was added, and mycobacterial cell suspension was transferred to microcentrifuge tube containing 0.1-mm-sized zirconia-silica bead (BioSpec, Oklahoma). Mycobacterium cells were disrupted three times (Speed No.2, 1 min) by OMNI Bead Ruptor (OMNI, USA). RNA isolation and cDNA conversion were further performed using reagents provided in the commercial kit. The amount of RNA and DNA was determined by measuring the optical density between 260 and 280 nm using a UV Spectrophotometer (Biotech Epoch™) and samples were kept at −20°C before use.
The Multiplex Real-Time PCR Assay Targets Stress-Related Genes
To determine the expression level of stress-related genes (hspX, tgs1, and sigE) of MTB induced by isoniazid, multiplex real-time PCR assay was designed and used in this work. Primers and probes were designed using NCBI blast and PrimerQuest tool except hspX. Primers and probes were listed in Table 1 and Table 2, respectively. The PCR reaction mixture (20 μL) consists of the following components: 1xTHUNDERBIRD™ Probe qPCR Mix (Toyobo, Japan), 0.3 µM of each primer, 0.25 µM of each probe, and 100 ng of cDNA template. Multiplex real-time PCR assay was performed using The CFX96 Touch Real-Time PCR Detection System (Bio-Rad, LABORATOIRES INC.). PCR conditions are shown in Table 3. The fold change of gene expression was calculated by the 2−ΔΔCT method.33 The housekeeping gene, sigA (Rv2703c)31 was used to normalize the expression levels of each interested gene. The PCR product was run on electrophoresis to confirm the size and specificity of the designed primers.
 |
Table 1 Primer Sequences for Multiplex Real-Time PCR Target Stress-Related Genes |
 |
Table 2 Probes Sequences Used in Multiplex Real-Time PCR Target Stress-Related Genes |
 |
Table 3 The Condition for Multiplex Real-Time PCR Assay Target Stress-Related Genes |
qRT-PCR Analysis Targets ManLAM-Related Genes
To determine the influence of INH stress on the regulation of ManLAM-related genes (pimB, mptA, mptC, dprE1, dprE2, and embC) in MTB, qRT-PCR was performed using cDNA of treated-MTB and untreated-MTB as a template. The specificity test of each primer was confirmed by PCR and agarose gel electrophoresis. Primer sequences are shown in Table 4, and PCR conditions are shown in Table 5. The PCR reaction mixture (20 μL) consists of the following components: 1xTHUNDERBIRD® SYBR® qPCR Mix (Toyobo, Japan), 0.4 µM of primers, and 100 ng of cDNA template. Real-time PCR was performed using The CFX96 Touch Real-Time PCR Detection System (Bio-Rad, LABORATOIRES INC.). The fold change of gene expression was calculated by the 2−ΔΔCT method.33 The housekeeping gene, sigA (Rv2703c)31 was used to normalize the expression levels of each interested gene.
 |
Table 4 Primer Sequences for ManLAM-Related Genes and Genes Associated with Drug Resistance |
 |
Table 5 PCR Conditions for ManLAM-Related Genes Expression Analysis |
Sequence Analysis
To study genetic variation of genes associated with drug resistance (inhA, katG, and rpoB) and ManLAM-related genes (pimB, mptA, mptC, dprE1, dprE2, and embC), DNA extraction and sanger sequencing were performed. MTB colonies were resuspended in 25 µL of 1 mg/mL lysozyme and incubated in water bath at 37°C for 10 min. Twenty-five microliters of proteinase K and 75 µL of 0.1 M Tris HCl were added and incubated in water bath at 37°C for 10 min. Then it was incubated in a hot block at 95°C for 10 min. The amount of DNA was determined by measuring the optical density of 260 and 280 nm using a UV Spectrophotometer (Biotech Epoch™) and kept at 4 or-20 °C before use. PCR was performed using Lab Cycler Basic-Thermal Cycler (SensoQuest, Germany) to amplify drug resistance-associated genes (inhA, katG, and rpoB) and ManLAM-related genes (pimB, mptA, mptC, dprE1, dprE2, and embC). Primer sequences are shown in Table 4 and PCR conditions are shown in Table 6. The PCR reaction mixture (50 μL) consists of the following components: 1xKOD OneTM PCR Master Mix (Toyobo, Japan), 0.3 µM of primers, and 100 ng of DNA template. After the PCR process, agarose gel electrophoresis was performed to investigate PCR product size. The single band as their expected product size without non-specific bands demonstrates the specificity of designed primers and appropriate PCR conditions. The NucleoSpin® Gel and PCR Clean-up (MACHEREY-NAGEL, Germany) were used to purify all PCR products and were measured by UV Spectrophotometer (Biotech Epoch™). For Sanger sequencing, purified DNA was mixed with BigDye™ Terminator v3.1 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, USA), primers (Table 4), and subjected to the automated sequencer ABI Prism 3730 XL. Sequence alignment was performed by BioEdit and NCBI nucleotide blast (BLASN) programs using Mycobacterium tuberculosis H37Rv (GenBank: CP003248.2) as a reference strain.
 |
Table 6 PCR Conditions for DNA Sequencing |
Statistical Analysis
The result was represented by the mean values of three independent experiments run in triplicate ± SEM (standard error means). Statistical analysis was performed in GraphPad Prism v8 using one-way analysis of variance with Tukey’s multiple comparison to assess the data between different groups. Kruskal–Wallis was used when data are not normally distributed. P-value <0.05 was considered statistically significant and * = P < 0.05, ** = P < 0.01, and *** = P < 0.001.
Results
Drug Resistance Profiles of MTB Samples
Drug-sensitivity test was investigated using the GenoType MTBDRplus assay and agar proportion method to detect the genotypic and phenotypic drug resistance profiles of MTB samples, respectively. In the GenoType MTBDRplus assay, RIF resistance was detected using a probe specific to rpoB gene, while INH resistance was detected using probes target katG and inhA genes. INH-R exhibited a mutation in the inhA regulatory region (C-15T), while RIF-R exhibited rpoB mutation (H526D). In MDR-MTB, double mutations of katG and rpoB were found at codon 315 (S315T) and 516 (D516V), respectively as presented in Figure S1 (Supplementary Materials). In the agar proportion method, mycobacteria were cultured with isoniazid, rifampin, streptomycin, ethambutol, and p-Nitrobenzoic acid. H37Rv was susceptible to all drugs, INH-R was resistant to isoniazid, RIF-R was resistant to rifampin, and MDR resistant to all drugs except p-Nitrobenzoic acid as presented in Table S1 (Supplementary Materials). These results correlate with the GenoType MTBDRplus assay.
The Expression of Stress-Related Genes in Response to INH Treatment
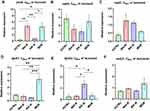
In this experiment, INH was used as a stressor and hspX, tgs1, and sigE were used as stress responder markers. H37Rv, INH-R and RIF-R and MDR were treated with INH at CMax concentration for 30 min to determine the effect of antibiotic stress. Results were presented as relative expression, which is the fold-changes of gene expression normalized to sigA expression relative to untreated control. Primers and probes for the multiplex real-time PCR assay used in this work were specific to targeted genes, and a non-specific band was not observed as presented in Figure S2 (Supplementary Materials). In response to INH, stress-related genes were regulated differently in each MTB strain. For hspX gene expression, 1.96 and 1.42-fold changes were observed in RIF-R and MDR, respectively (Figure 1A). Interestingly, tgs1 was markedly up-regulated in MDR (2.03-fold) and significantly higher than others (Figure 1B). However, H37Rv and RIF-R also up-regulated tgs1 at 1.26 and 1.09-fold, respectively (Figure 1B). Surprisingly, H37Rv (drug-sensitive strain) was clearly up-regulated sigE (3.07-fold) among four strains (Figure 1C). The findings indicate that INH treatment can cause stress in mycobacteria, resulting in increased expression of stress responder genes. The expression level varied between MTB strains with distinct drug resistance profiles, highlighting the unique potential of their adaptation ability. Additionally, MDR-MTB was more strongly responsive to INH stress than other strains, implying a relationship between the stress response mechanism and drug resistance.
The Expression Level of ManLAM-Related Genes in Response to INH Treatment
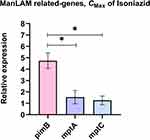
The specificity of primers to targeted genes was confirmed using MTB samples including H37Rv (a drug-sensitive strain), INH-R, RIF-R, and MDR. All primers tested produced a single band at a desired product size, demonstrating primer specificity as presented in Figure S3 (Supplementary Materials). After INH treatment, the relative expression of ManLAM-related genes was determined for each MTB strain, and untreated MTB was used as a control. Results were presented as relative expression, compared between four strains (Figure 2). In H37Rv, only mptA that involved in the elongation of LM was up-regulated (2.84-fold), but the relative expression was not significantly different among the four strains (Figure 2B). Interestingly, in INH-R, pimB, mptA, and mptC were up-regulated. pimB is one of the genes responsible for PIMs production, which is involved in an early stage of ManLAM biosynthesis. PimB was up-regulated 4.73-fold and expressed significantly higher than H37Rv and RIF-R (p<0.01) (Figure 2A). mptA and mptC that are involved in elongation and branching of LM/LAM were up-regulated 1.54 and 1.26-fold, respectively (Figure 2B and C). In RIF-R, ManLAM-related genes were slightly up-regulated, including mptA (1.24-fold), mptC (1.12-fold), and dprE2 (1.37-fold) (Figure 2B, C, and E). In MDR, several genes were markedly up-regulated, including pimB (4.55-fold), mptA (2.48-fold) and dprE1 (3.2-fold) (Figure 2A, B, D). MDR up-regulated pimB greater than H37Rv and RIF-R (p<0.05), while predominantly up-regulated dprE1 among four strains. However, embC that involved in the biosynthesis of branched arabinan polymers on arabinogalactan was also upregulated in MDR by 1.37-fold, but not statistically significantly higher than others (Figure 2F). Among ManLAM-related genes that were up-regulated in each strain, pimB was strongly up-regulated greater than mptA and mptC in INH-R (Figure 3). Following experimental results, indicate that the expression pattern was significantly different between drug-sensitive (H37Rv) and drug-resistant TB (Figure 4). These results suggest that INH treatment induce ManLAM-related genes expression, especially in drug-resistant TB.
Sequence Analysis of Drug Resistance-Associated Genes
Sequence analysis was performed to study the genetic variation of drug resistance-associated genes in MTB used in this work. INH-R MTB exhibited a C-15T mutation in the inhA promoter region (Figure 5). In RIF-R, rpoB mutation (H526D) was observed (Figure 6). MDR exhibited both mutations, including rpoB mutation (D516V) (Figure 6) and katG mutation (S513T) (Figure 7). These findings were similar to results from the GenoType MTBDRplus assay.
 |
Figure 5 Sequence alignment of inhA for MTB with different drug resistance profiles. *Represents the position of the nucleotide mutation in the inhA regulatory region of INH-R. |
Sequence Analysis of ManLAM-Related Genes
No differences in the DNA sequence of ManLAM-related genes were observed, except for the dprE1 (Rv3790) gene. In RIF-R and MDR isolates, C→T mutation at nucleotide position 459 of dprE1 (Rv3790) was observed, resulting in the same amino acid at codon position 153 (Figure 8). This mutation point was matched with 96 samples from the NCBI database, especially in drug-resistant MTB. These MTB samples are mostly found in India (28%) and China (13%). However, drug-sensitive MTB also exhibited a dprE1 (Rv3790) mutation at the same position, indicating that this mutation is a genetic polymorphism and may be unrelated to drug resistance.
 |
Figure 8 Sequence alignment of dprE1 for MTB with different drug resistance profiles. *Indicates the location of the nucleotide mutation in dprE1 gene of MDR and RIF-R. |
Discussion
Several publications have reported that MTB responds to several stresses by remodeling cell wall components to promote their survival in different conditions, and these changes also affect their antibiotic susceptibility profiles.3,34,35 The model of cell wall regulation from a recent review article demonstrates that LM and LAM are moderately expressed in rapid growth but up-regulated during growth stasis.36 Changes in LM and LAM structures significantly impact the cell wall integrity of M. smegmatis, resulting in loss of the acid-fast staining property, increased sensitivity to antibiotics, and more sensitivity to macrophage killing. Similarly, defects in LM and LAM structures affect the pathogenesis of MTB.37 Thus, these suggest that MTB regulates ManLAM expression, which is a virulence factor for protecting mycobacterial cells and supporting their long-term survival.
Previous publication categorized hspX and tgs1 as dormancy regulon while sigE was classified as an enduring hypoxic response according to their function or expression pattern.18 Both hspX and tgs1 genes were clearly up-regulated in Wayne dormancy culture model (hypoxia). sigE was also up-regulated in this model but presented at a lower level compare to hspX and tgs1 at the indicated time point. Similar result was observed, hspX and tgs1 were significantly expressed in multi-stress model3 [low oxygen, higher levels of carbon dioxide, nutrient starvation, and acidic pH (5.0)]. This multi-stress model induces MTB switch from an active to a dormancy phase, resulting in long-term survival and drug tolerance. Thus, these data suggest that environmental stress triggers MTB to regulate stress responder genes to promote their survival. A similar phenomenon may occur in the case of antibiotic stress such as INH that is shown in the up-regulation of hspX, tgs1 and sigE in this work.
This study was interested in the stress response mechanism of M. tuberculosis induced by INH drug. The aim of this study was to determine the expression level of ManLAM-related genes in response to INH in M. tuberculosis clinical isolates with various drug resistance profiles. Interestingly, ManLAM-related genes tested in this study were regulated differently between each drug-resistant strain, which causes a unique pattern. Almost all ManLAM-related genes were up-regulated in drug-resistant strains, especially in MDR.
MDR isolates have katG (S513T) and rpoB (D516V) mutations, which are associated with INH resistance and RIF resistance, respectively. Even though INH-R and RIF-R present one phenotypic drug-resistant profile as present in MDR, the pattern of ManLAM-related gene expression was significantly different. INH-R has C-15T mutation in the inhA promoter region, while RIF-R has rpoB mutation (H526D). These results suggest that the genetic background is one of the factors involved in the adaptive response mechanism of MTB under INH stress.
Interestingly, INH-R and MDR were dramatically up-regulated pimB. pimB is one of the genes responsible for PIMs synthesis, the structural basis of ManLAM. PIMs play a role in maintaining cell wall integrity that provides mycobacteria with intrinsic antibiotic tolerance and modulates host response during infection.34 PIMs activate cytokine production that induces granuloma formation and promotes disease progression.38,39 The expression level of mptA was up-regulated in all strains at similar levels, reflecting its requirement under INH stress conditions (Figure 2B). The balance activity of mptA and mptC is crucial for LM/LAM production in mycobacteria. Overexpression of MSMEG_4247, which is an orthologs of Rv2181 (mptC) leads to the synthesis of dwarfed LM, shorter mannan backbone and smaller arabinan domain of LAM in M. smegmatis.37 The expression level of mptA and mptC following INH treatment in this study was not significantly different in each strain except H37Rv (drug-sensitive strain), suggesting that the balance of these two genes’ expression also plays a role in the adaptive mechanism of MTB. The expression level of dprE1 and dprE2 was up-regulated in MDR and RIF-R, respectively. dprE1 and dprE2 are responsible for the epimerization of DPR to DPA,40 arabinose donor in the arabinan domains of arabinogalactan and ManLAM. DprE1 is essential in mycobacteria and it is a valuable drug target.41 dprE1 depletion induces cell lysis, which results in cell death and is attenuated in macrophages.42 DprE1 mutation affect mycobacterial growth and virulence, such as levels of resistance to drugs and changes in infectivity, depend on mutation type and position.43 Alignment of RIF-R and MDR in this study showed a mutation at nucleotide position 459 of the dprE1 gene that presents the same amino acid at codon 153. However, dprE1 up-regulation was observed only in MDR, not RIF-R, suggesting that there are other factors that regulate dprE1 expression response to INH treatment. embC (Rv3793) was up-regulated only in MDR. Previous work reported that embC is essential in MTB and the expression level of embC influenced the size of LAM in M. smegmatis but not in MTB. Moreover, the embC promoter was down-regulated under the hypoxia-induced nonreplicating condition, and embC was down-regulated in alveolar macrophages at an early stage of infection.12 Thus, suggest that embC is regulated by mycobacteria depending on the type and duration of stressors.
Conclusion
The adaptive mechanism of mycobacteria in response to stressors is an important strategy to promote their virulence and pathogenesis. This study determined the effect of antibiotic stress on Mycobacterium tuberculosis (MTB) by focusing on mannose-capped lipoarabinomannan (ManLAM), which is one of the virulence factors that modulates host immune response. Multiplex real-time PCR assay targets stress responder genes and qRT-PCR targets ManLAM-related genes were performed. Isoniazid acts as a stressor to induce stress response in mycobacteria, as shown in the up-regulation of stress-related genes including hspX, tgs1, and sigE. The expression pattern of ManLAM-related genes in drug resistance and drug-sensitive MTB in response to INH was different, causing a unique pattern. ManLAM-related genes respond to isoniazid mostly in drug-resistant strains and are present at high expression levels in INH-R and MDR. The results suggest that ManLAM is one factor involved in the adaptive mechanism of MTB response to isoniazid stress. This work provides new insights into mycobacterial adaptation response to isoniazid that will improve our understanding of how mycobacteria develop drug resistance in the future.
Acknowledgments
This project is funded by the National Research Council of Thailand (NRCT) [NRCT5-RSA63004-14], Thailand Research Fund (TRF) through the Royal Golden Jubilee Ph.D. Program (RGJ PhD) [PHD/0114/2561] and CMU research fellowship program. The authors would like to thank Faculty of Associated Medical Sciences, Chiang Mai University for their support with the equipment.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Nasiri MJ, Haeili M, Ghazi M, et al. New insights in to the intrinsic and acquired drug resistance mechanisms in mycobacteria. Front Microbiol. 2017;8:681. doi:10.3389/fmicb.2017.00681
2. Nguyen L. Antibiotic resistance mechanisms in M. tuberculosis: an update. Arch Toxicol. 2016;90(7):1585–1604. doi:10.1007/s00204-016-1727-6
3. Deb C, Lee CM, Dubey VS, et al. A novel in vitro multiple-stress dormancy model for Mycobacterium tuberculosis generates a lipid-loaded, drug-tolerant, dormant pathogen. PLoS One. 2009;4(6):e6077. doi:10.1371/journal.pone.0006077
4. Jarlier V, Nikaido H. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol Lett. 1994;123(1–2):11–18. doi:10.1111/j.1574-6968.1994.tb07194.x
5. Rajni RN, Meena LS. Biosynthesis and virulent behavior of lipids produced by Mycobacterium tuberculosis: LAM and cord factor: an overview. Biotechnol Res Int. 2011;2011:274693. doi:10.4061/2011/274693
6. Zhou KL, Li X, Zhang XL, et al. Mycobacterial mannose-capped lipoarabinomannan: a modulator bridging innate and adaptive immunity. Emerg Microbes Infect. 2019;8(1):1168–1177. doi:10.1080/22221751.2019.1649097
7. Mishra AK, Driessen NN, Appelmelk BJ, et al. Lipoarabinomannan and related glycoconjugates: structure, biogenesis and role in Mycobacterium tuberculosis physiology and host-pathogen interaction. FEMS Microbiol Rev. 2011;35(6):1126–1157. doi:10.1111/j.1574-6976.2011.00276.x
8. Mishra AK, Batt S, Krumbach K, et al. Characterization of the Corynebacterium glutamicum deltapimB’ deltamgtA double deletion mutant and the role of Mycobacterium tuberculosis orthologues Rv2188c and Rv0557 in glycolipid biosynthesis. J Bacteriol. 2009;191(13):4465–4472. doi:10.1128/JB.01729-08
9. Mishra AK, Alderwick LJ, Rittmann D, et al. Identification of an alpha (1–>6) mannopyranosyltransferase (MptA), involved in Corynebacterium glutamicum lipomanann biosynthesis, and identification of its orthologue in Mycobacterium tuberculosis. Mol Microbiol. 2007;65(6):1503–1517. doi:10.1111/j.1365-2958.2007.05884.x
10. Kaur D, Berg S, Dinadayala P, et al. Biosynthesis of mycobacterial lipoarabinomannan: role of a branching mannosyltransferase. Proc Natl Acad Sci U S A. 2006;103(37):13664–13669. doi:10.1073/pnas.0603049103
11. Angala SK, Belardinelli JM, Huc-Claustre E, et al. The cell envelope glycoconjugates of Mycobacterium tuberculosis. Crit Rev Biochem Mol Biol. 2014;49(5):361–399. doi:10.3109/10409238.2014.925420
12. Goude R, Amin AG, Chatterjee D, et al. The critical role of embC in Mycobacterium tuberculosis. J Bacteriol. 2008;190(12):4335–4341. doi:10.1128/JB.01825-07
13. Kaur D, Obregón-Henao A, Pham H, et al. Lipoarabinomannan of Mycobacterium: mannose capping by a multifunctional terminal mannosyltransferase. Proc Natl Acad Sci U S A. 2008;105(46):17973–17977. doi:10.1073/pnas.0807761105
14. Liu Y, Tan S, Huang L, et al. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J Exp Med. 2016;213(5):809–825. doi:10.1084/jem.20151248
15. Magombedze G, Dowdy D, Mulder N. Latent tuberculosis: models, computational efforts and the pathogen’s regulatory mechanisms during dormancy. Front Bioeng Biotechnol. 2013;1:4. doi:10.3389/fbioe.2013.00004
16. Stewart GR, Newton SM, Wilkinson KA, et al. The stress-responsive chaperone alpha-crystallin 2 is required for pathogenesis of Mycobacterium tuberculosis. Mol Microbiol. 2005;55(4):1127–1137. doi:10.1111/j.1365-2958.2004.04450.x
17. Sirakova TD, Dubey VS, Deb C, et al. Identification of a diacylglycerol acyltransferase gene involved in accumulation of triacylglycerol in Mycobacterium tuberculosis under stress. Microbiology. 2006;152(Pt 9):2717–2725. doi:10.1099/mic.0.28993-0
18. Iona E, Pardini M, Mustazzolu A, et al. Mycobacterium tuberculosis gene expression at different stages of hypoxia-induced dormancy and upon resuscitation. J Microbiol. 2016;54(8):565–572. doi:10.1007/s12275-016-6150-4
19. Veatch AV, Niu T, Caskey J, et al. Sequencing-relative to hybridization-based transcriptomics approaches better define Mycobacterium tuberculosis stress-response regulons. Tuberculosis. 2016;101S:S9–S17. doi:10.1016/j.tube.2016.09.020
20. Hampshire T, Soneji S, Bacon J, et al. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis. 2004;84(3–4):228–238. doi:10.1016/j.tube.2003.12.010
21. Baker JJ, Dechow SJ, Abramovitch RB. Acid fasting: modulation of Mycobacterium tuberculosis metabolism at Acidic pH. Trends Microbiol. 2019;27(11):942–953. doi:10.1016/j.tim.2019.06.005
22. Bansal R, Anil Kumar V, Sevalkar RR, et al. Mycobacterium tuberculosis virulence-regulator PhoP interacts with alternative sigma factor SigE during acid-stress response. Mol Microbiol. 2017;104(3):400–411. doi:10.1111/mmi.13635
23. Deretic V, Pagán-Ramos E, Zhang Y, et al. The extreme sensitivity of Mycobacterium tuberculosis to the front-line antituberculosis drug isoniazid. Nat Biotechnol. 1996;14(11):1557–1561. doi:10.1038/nbt1196-1557
24. Vilchèze C, Jacobs WR
25. Palomino JC, Martin A. Drug resistance mechanisms in Mycobacterium tuberculosis. Antibiotics. 2014;3(3):317–340. doi:10.3390/antibiotics3030317
26. Wilson M, DeRisi J, Kristensen -H-H, et al. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc Natl Acad Sci U S A. 1999;96(22):12833–12838. doi:10.1073/pnas.96.22.12833
27. Vilcheze C, Jacobs WR
28. Nieto RL, Mehaffy C, Islam MN, et al. Biochemical characterization of isoniazid-resistant Mycobacterium tuberculosis: can the analysis of clonal strains reveal novel targetable pathways? Mol Cell Proteomics. 2018;17(9):1685–1701. doi:10.1074/mcp.RA118.000821
29. Boot M, van Winden VJC, Sparrius M, et al. Cell envelope stress in mycobacteria is regulated by the novel signal transduction ATPase IniR in response to trehalose. PLoS Genet. 2017;13(12):e1007131. doi:10.1371/journal.pgen.1007131
30. Alsultan A, Peloquin CA. Therapeutic drug monitoring in the treatment of tuberculosis: an update. Drugs. 2014;74(8):839–854. doi:10.1007/s40265-014-0222-8
31. Gamngoen R, Putim C, Salee P, Phunpae P, Butr-Indr B. A comparison of Rv0559c and Rv0560c expression in drug-resistant Mycobacterium tuberculosis in response to first-line antituberculosis drugs. Tuberculosis. 2018;108:64–69. doi:10.1016/j.tube.2017.11.002
32. Woods GL, Brown-Elliott BA, Conville PS, et al. Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes.
33. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
34. Queiroz A, Riley LW. Bacterial immunostat: Mycobacterium tuberculosis lipids and their role in the host immune response. Rev Soc Bras Med Trop. 2017;50(1):9–18. doi:10.1590/0037-8682-0230-2016
35. Cunningham AF, Spreadbury CL. Mycobacterial stationary phase induced by low oxygen tension: cell wall thickening and localization of the 16-kilodalton alpha-crystallin homolog. J Bacteriol. 1998;180(4):801–808. doi:10.1128/JB.180.4.801-808.1998
36. Dulberger CL, Rubin EJ, Boutte CC. The mycobacterial cell envelope - a moving target. Nat Rev Microbiol. 2020;18(1):47–59. doi:10.1038/s41579-019-0273-7
37. Sena CBC, Fukuda T, Miyanagi K, et al. Controlled expression of branch-forming mannosyltransferase is critical for mycobacterial lipoarabinomannan biosynthesis. J Biol Chem. 2010;285(18):13326–13336. doi:10.1074/jbc.M109.077297
38. Koul A, Herget T, Klebl B, et al. Interplay between mycobacteria and host signalling pathways. Nat Rev Microbiol. 2004;2(3):189–202. doi:10.1038/nrmicro840
39. Nigou J, Vasselon T, Ray A, et al. Mannan chain length controls lipoglycans signaling via and binding to TLR2. J Immunol. 2008;180(10):6696–6702. doi:10.4049/jimmunol.180.10.6696
40. Bhutani I, Loharch S, Gupta P, et al. Structure, dynamics, and interaction of Mycobacterium tuberculosis (Mtb) DprE1 and DprE2 examined by molecular modeling, simulation, and electrostatic studies. PLoS One. 2015;10(3):e0119771. doi:10.1371/journal.pone.0119771
41. Crellin PK, Brammananth R, Coppel RL. Decaprenylphosphoryl-β-D-ribose 2′-epimerase, the target of benzothiazinones and dinitrobenzamides, is an essential enzyme in Mycobacterium smegmatis. PLoS One. 2011;6(2):e16869. doi:10.1371/journal.pone.0016869
42. Kolly GS, Boldrin F, Sala C, et al. Assessing the essentiality of the decaprenyl-phospho-d-arabinofuranose pathway in Mycobacterium tuberculosis using conditional mutants. Mol Microbiol. 2014;92(1):194–211. doi:10.1111/mmi.12546
43. Foo CS-Y, Lechartier B, Kolly GS, et al. Characterization of DprE1-mediated benzothiazinone resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2016;60(11):6451–6459. doi:10.1128/AAC.01523-16
44. Schaeffer ML, Khoo KH, Besra GS, et al. The pimB gene of Mycobacterium tuberculosis encodes a mannosyltransferase involved in lipoarabinomannan biosynthesis. J Biol Chem. 1999;274(44):31625–31631. doi:10.1074/jbc.274.44.31625
45. Korkegian A, Roberts DM, Blair R, et al. Mutations in the essential arabinosyltransferase embc lead to alterations in Mycobacterium tuberculosis lipoarabinomannan. J Biol Chem. 2014;289(51):35172–35181. doi:10.1074/jbc.M114.583112
46. Mikusová K, Huang H, Yagi T, et al. Decaprenylphosphoryl arabinofuranose, the donor of the D-arabinofuranosyl residues of mycobacterial arabinan, is formed via a two-step epimerization of decaprenylphosphoryl ribose. J Bacteriol. 2005;187(23):8020–8025. doi:10.1128/JB.187.23.8020-8025.2005
47. Ramirez MV, Cowart KC, Campbell PJ, et al. Rapid detection of multidrug-resistant Mycobacterium tuberculosis by use of real-time PCR and high-resolution melt analysis. J Clin Microbiol. 2010;48(11):4003–4009. doi:10.1128/JCM.00812-10
48. Chikaonda T, Ketseoglou I, Nguluwe N, et al. Molecular characterisation of rifampicin-resistant Mycobacterium tuberculosis strains from Malawi. Afr J Lab Med. 2017;6(2):463. doi:10.4102/ajlm.v6i2.463
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.