Back to Journals » OncoTargets and Therapy » Volume 10
The prognostic value of decreased miR-101 in various cancers: a meta-analysis of 12 studies
Authors Hu J, Wu C, Zhao X, Liu C
Received 12 May 2017
Accepted for publication 25 June 2017
Published 24 July 2017 Volume 2017:10 Pages 3709—3718
DOI https://doi.org/10.2147/OTT.S141652
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr XuYu Yang
Jianpei Hu, Chunyu Wu, Xueying Zhao, Chaodong Liu
Department of Urology, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
Background: A consensus regarding the prognostic value of decreased miR-101 in human cancers has not been reached. This study aimed to comprehensively investigate the internal associations between loss of miR-101 expression and prognostic implications in patients with cancer.
Materials and methods: All relevant literature in electronic databases, including PubMed, ISI Web of Science, and Embase, up to March 1, 2017 were searched. Correlations between decreased miR-101 and clinicopathological parameters were defined by odds ratios (ORs). The degree of association between reduced miR-101 and survival outcome was evaluated by pooled hazard ratios (HRs) and relevant 95% CIs.
Results: Twelve eligible studies with 2,088 patients were included in this meta-analysis. Decreased miR-101 expression was closely connected with poor overall survival, with a pooled HR of 2.15 (95% CI 1.71–2.7, P<0.001). This correlation was also revealed when stratified analysis was conducted with respect to ethnicity, cancer type, sample size, specimen source, and analysis model. However, decreased miR-101 was not associated with disease-free survival, recurrence-free survival, or progression-free survival, with a pooled HR of 1.59 (95% CI 0.83–3.03, P=0.128), despite a positive trend. In addition, reduced miR-101 was intimately related to poorer tumor differentiation (OR 2.17, 95% CI 1.14–4.13; P=0.019), advanced tumor classification (OR 5.25, 95% CI 3.39–8.12; P<0.001), and higher TNM stage (OR 6.18, 95% CI 3.79–10.09; P<0.001).
Conclusion: Our findings suggest that loss of miR-101 expression is correlated with worse overall survival in a variety of cancers, and could serve as a predictive indicator for clinicopathological features. Furthermore, miR-101 may become a feasible therapeutic target in most human cancers.
Keywords: cancer, miR-101, prognosis, meta-analysis
Introduction
In the last decade, primarily through early detection or diagnosis and improved treatment progress, the overall death rate of cancer has continuously declined by approximately 1.5% each year in the US.1 However, the extent of this decline is unsatisfactory, and cancer remains the leading cause of death throughout the world, suggesting that continuous efforts toward cancer research are warranted. To date, despite cancer remaining an incurable disease, survival and quality of life in cancer patients have improved. This is at least partly contributed to by the reasonable prognosis in these patients, leading to more precise risk stratification, as well as optimal choice of treatment.2 Therefore, the identification of novel biological tumor makers that could supplement the current TNM stage-based predictive system can thereby assist clinicians in improving treatments in a clinical setting.
miRNAs are members of the non-protein-coding RNA family, and are small endogenous RNA molecules around 22 nucleotides in length that are highly conserved in mammalian evolution. After binding to complementary sequences, miRNAs can degrade or silence target mRNAs posttranscriptionally.3,4 In particular, published studies have demonstrated that dysregulated miRNA patterns (upregulation and/or downregulation) contribute to tumor initiation and progression in various malignancies via the modulation of target genes involved in proliferation, apoptosis, angiogenesis, invasion, and migration.5,6 In addition, from a clinical practice perspective, the value of miRNAs as diagnostic and prognostic markers has been revealed in recent years.7,8
As one of the most studied miRNAs, miR-101 plays a crucial role in cancer biology through its inhibition of various cellular processes, including proliferation, chemoresistance, angiogenesis, invasion, and metastasis,9,10 suggesting that it could be a tumor-suppressor miRNA. In line with this, multiple studies have reported that miR-101 undergoes downregulation in multiple cancers and that loss of miR-101 expression is linked to unfavorable prognosis in cancers, such as non-small-cell lung cancer (NSCLC),11,12 gallbladder cancer,13 breast cancer,14 glioma,15,16 bladder cancer,17 laryngeal squamous-cell carcinoma,18 hepatocellular carcinoma (HCC),19,20 and colorectal cancer (CRC).21 However, the predictive value of miR-101 remains inconclusive, due to the existence of opposing results. For instance, Lv et al reported that miR-101 was upregulated in HCC and correlated with worse survival outcomes for patients.22 In summary, there is not enough evidence to draw conclusions about the prognostic value of miR-101 in malignancies, due to these contradictory results. Therefore, a comprehensive meta-analysis based on a larger sample size was conducted to clarify the value of miR-101 as a predictive biomarker for patients’ clinical outcomes by analyzing published studies concerning a variety of cancer types.
Materials and methods
Search strategy
This meta-analysis was conducted by complying with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines.23 To obtain related articles, thorough bibliographic retrieval was conducted in the electronic databanks PubMed, Embase, and ISI Web of Science. By use of relevant synonyms and Boolean logic, exhaustive searches were run with the search details (microRNA-101 OR miR-101 OR mir101) AND (cancer OR tumor OR neoplasm OR carcinoma) AND (prognosis OR prognostic OR survival OR outcome). The search was refreshed up to March 1, 2017 without a lower date limit. Additionally, reference lists of identified studies were further assessed manually to find potential missing studies.
Selection criteria
Studies were considered on the proviso that they met all of the following criteria: 1) research subjects were restricted to humans, 2) miR-101 expression was detected with quantitative real-time polymerase chain reaction (qRT-PCR) or in situ hybridization in cases with any confirmed type of solid tumor, 3) the interrelation of miR-101 expression level and patients’ survival outcome was revealed explicitly and/or by using clinicopathological parameters, including tumor differentiation, tumor classification, lymph-node classification, and TNM stage, 4) hazard ratios (HRs) and 95% CIs were provided firsthand or indirectly through papers (Kaplan–Meier curves) or authors (original survival data), and 5) the available full text was written in English.
Studies were removed if they met any of the following criteria: 1) nonhuman studies, 2) improper article types, such as reviews, meeting abstracts, comments, and case reports, 3) the available information was not enough to calculate HRs and 95% CIs, and 4) the articles were in languages other than English. Only the most complete study was selected when two or more studies reported their results based on the same population. This was assessed by two reviewers (JH and CW), and different opinions were assessed by discussion with a third reviewer (XZ).
Data extraction and quality assessment
Collected data were first author, year of publication, country of origin, tumor type, clinicopathological features, number of included cases, source of specimens, method of detection, cutoff value for categorizing miR-101-expression level, end point, time of follow-up visits, HRs and 95% CIs, and the source of HRs (univariate or multivariate analysis). Where both uni- and multivariate analyses were given, the latter was adopted for its higher reliability. Additionally, if direct HRs and 95% CIs could not be obtained from the studies, Engauge Digitizer 4.1 software was utilized to extract patients’ survival data from Kaplan–Meier survival curves and a method reported by Tierney et al24 was used to calculate indirect HRs with 95% CIs.
Quality assessment of identified studies was carried out based on the Newcastle–Ottawa scale, which is recommended in meta-analysis for observational and nonrandomized studies.25 Three distinct aspects – selection, comparability, and outcome – constitute this widely used scale. Through comprehensive assessment of each item, final stars were given to each publication. Generally, one study could be awarded zero to nine stars, and studies possessing more than five stars were considered high quality in methodology and included for further analyses. Two authors (JH and XZ) separately performed these steps, after which a cross-check was done and any discrepancies resolved by discussion.
Statistical analysis
Correlations between decreased miR-101 and clinicopathological parameters were defined by odds ratios (ORs). In general, an OR >1 indicated that loss of miR-101 expression was more likely to correlate with poorer tumor-differentiation degree and a higher TNM stage.
In the meta-analysis presented here, in view of all of the literature analyzed for time-to-survival data, HR and CI estimation (95%) was designated the effect magnitude, in order to interpret the impact of miR-101 absence on patient prognosis. HRs and 95% CIs derived from each individual paper were synthesized for an integrated and more accurate result. By convention, a combined HR >1 predicted that decreased expression of miR-101 was associated with an unfavorable survival outcome, and differences between groups were statistically significant when estimated 95% CIs did not contain 1. Heterogeneity across included publications was inspected qualitatively and quantitatively via Q-test (Cochrane Handbook recommended) and Higgins’s I2,26 respectively. Unacceptable heterogeneity was defined as P<0.05 and/or I2>50%, resulting in the use of a random-effect model for the estimation on summary statistics. In contrast, the fixed-effect model was taken to assess the combined effect size. Publication bias from analytical studies was checked by virtue of Begg funnel plots27 together with Egger’s method.28 Generally, no obvious asymmetry from funnel plots and P>0.05 from Egger’s test were considered an absence of potential publication bias. Furthermore, sensitivity analysis for evaluation of data robustness was conducted by removing each study one by one to examine the impact of a single study on the overall results. Two-sided P<0.05 possessed statistical significance. All analyses were performed with Stata version 13.0 (Stata Corp, College Station, TX, USA).
Results
Search results
A total of 553 publications were retrieved from the initial search of online databases, and 406 distinct studies remained after duplicates were removed. Through careful reading of titles and abstracts, 380 irrelevant studies were excluded, because they were not original studies (reviews, meeting abstracts, comments, and case reports), were not performed in humans, were not cancer-related studies, or were not focused on patient prognosis, and 26 studies with potential relevance were obtained and further assessed for eligibility based on a close reading of the full text. Following this, 12 eligible studies were finally included in the present analysis, and 14 studies were excluded due to irrelevance to prognosis (three), insufficient data to estimate HR (nine), combination with other biomarkers to predict prognosis (one), and unreliable prognostic estimation (HR 189.5 for overall survival [OS], n=1) on the basis of limited sample size (n=21). Figure 1 concisely summarizes the selection procedure of all eligible studies in our meta-analysis.
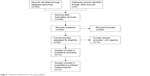  | Figure 1 Selection flowchart for the meta-analysis. |
Characteristics of included studies
The main characteristics of the 12 identified papers with 2,088 patients investigating the correlation between reduced miR-101 expression and clinical prognosis are shown in Table 1. As for the origin of the population, the vast majority of the included studies were carried out in China (eleven) and one study was from the US. In terms of cancer type among the identified studies, three papers focused on HCC, two on NSCLC, two on glioblastoma (GB), and several on other sporadic types, including gallbladder cancer, breast cancer, bladder cancer, laryngeal squamous-cell carcinoma, and CRC. Sample size differed among the studies, ranging from 42 to 1,134. With respect to the detection method used, eleven papers utilized qRT-PCR to determine miR-101 expression, while one study used in situ hybridization. Different cutoff values were utilized in those studies, where half used median value as the cutoff point. In total, three studies provided direct HRs with 95% CIs, while the remaining nine studies provided sufficient data to compute indirect HRs along with corresponding 95% CIs. A total of 12 studies analyzed the relation between decreased miR-101 expression and OS, whereas four studies determined the prognostic value of miR-101 deficiency for disease-free survival (DFS)/recurrence-free survival (RFS)/progression-free survival (PFS). Other detailed features were recorded, and are summarized in Table 1.
Association of decreased miR-101 with clinicopathological features
Main results regarding the impact of reduced miR-101 on clinicopathological features are shown in Table 2. Overall, eight studies focused on the association between decreased miR-101 and tumor-differentiation degree, demonstrating that reduced miR-101 expression was tightly connected with poorer tumor differentiation, with a pooled OR of 2.17 (95% CI 1.14–4.13, P=0.019). Furthermore, miR-101 deficiency was also related to tumor classification and TNM stage, with pooled ORs of 5.25 (95% CI 3.39–8.12, P<0.001) and 6.18 (95% CI 3.79–10.09, P<0.001), respectively. However, reduced miR-101 was not significantly related to lymph-node classification, with a pooled OR of 2.42 (95% CI 0.56–10.49, P=0.239), though the trend remained positive.
  | Table 2 Meta-analysis results of associations of decreased miR-101 expression with clinicopathological parameters |
Decreased miR-101 and OS
The association between decreased miR-101 in the OS prognosis of cancer patients is illustrated in Figure 2 on the basis of 12 studies with 2,088 patients. Overall, loss of miR-101 expression was closely linked to worse outcome for OS, with a pooled HR of 2.15 (95% CI 1.71–2.7, P<0.001), which was calculated from a fixed-effect model for insignificant heterogeneity among included articles (I2=41.2%, P=0.067).
  | Figure 2 Forest plot of studies evaluating HRs of decreased miR-101 expression for OS. |
Subgroup analysis exploring the source of heterogeneity was conducted for OS in accordance with ethnicity, specimen source, detection assay, tumor type, sample size, estimated HR, and analysis type, which is shown in Table 3. On the basis of ethnicity, the predictive value of decreased miR-101 did not change significantly, with combined HRs of 2.04 (95% CI 1.6–2.59, P<0.001) for Asians and 3.51 (95% CI 1.72–7.15, P<0.001) for non-Asians. When grouped based on specimen source, both subgroups exhibited associations where decreased miR-101 was an unfavorable indicator, with combined HRs of 1.97 (95% CI 1.55–2.52, P<0.001) for tissue samples and 3.91 (95% CI 2.06–7.42, P<0.001) for plasma samples. In the subgroup analysis by detection method, loss of miR-101 was related to poorer OS in the qRT-PCR group, with a pooled HR of 2.17 (95% CI 1.72–2.72, P<0.001). When stratifying by cancer type, miR-101 absence predicted a bad outcome in NSCLC (HR 1.69, 95% CI 1.01–2.83, P=0.044) and HCC (HR 2.18, 95% CI 1.41–3.37, P<0.001), but not in GB (HR 1.63, 95% CI 0.94–2.82; P=0.083). Although decreased miR-101 was not connected to worse OS for GB, the trend was positive. This statistical insignificance may have been due to limited studies (two) and low number of patients enrolled. Notably, in the subgroups of sample sizes >100, multivariate analysis and estimated HRs reported directly that loss of miR-101 expression demonstrated less heterogeneity and relatively stronger associations, with combined HRs of 2.65 (95% CI 1.93–3.65, P<0.001), 2.17 (95% CI 1.32–3.35, P<0.001), and 2.45 (95% CI 1.66–3.62, P<0.001), respectively.
Decreased miR-101 and DFS/RFS/PFS
As shown in Figure 3, four studies involving 389 patients revealed that decreased miR-101 did not correlate with DFS/RFS/PFS, with a pooled HR of 1.59 (95% CI 0.83–3.03, P=0.128) and estimated by a random-effect model for significant between-study heterogeneity (I2=72.7%, P=0.012); however, the trend was positive.
Publication bias and sensitivity analysis
Considering the limited number of included studies, only publication bias and sensitivity analysis for OS was performed. Begg funnel plots were used for evaluation of potential publication bias qualitatively via visual inspection. As shown in Figure 4, no obvious asymmetry was observed, together with the result of Egger’s tests assessing publication bias quantitatively (P=0.637), suggesting that the OS results were free of publication bias. In order to examine the reliability of the OS results, a sensitivity analysis was run, characterized by deletion of each study one by one. As shown in Figure 5, none of the omissions of individual articles changed the overall result significantly, which suggested strong robustness for the results.
  | Figure 4 Begg funnel plot for publication-bias test of overall survival. |
  | Figure 5 Sensitivity analysis of relationship between decreased miR-101 expression and overall survival in cancers. |
Discussion
miRNAs belong to a small noncoding RNA family (<200 nucleotides), which also contains several other subgroups classified by their diverse biological effects, such as snoRNAs, snRNAs, and siRNAs. Due to enormous advancements in experimental techniques, a set of feasible methods for miRNA detection have been well established, including reverse-transcriptase qRT-PCR, PCR-based arrays, and next-generation sequencing. Through this, researchers have identified increasing numbers of miRNAs.29 In recent years, studies have revealed the multiple roles played by miRNAs in physiological conditions and disease states, especially in cancers. Dysregulated miRNAs, involving downregulation of tumor-suppressor miRNAs and upregulation of oncogenic miRNAs, have been widely linked to tumor initiation, together with progression.30 To date, many miRNA signatures in various tumors have been correlated with diagnostic, prognostic, and therapeutic values.31 Of note, the utilization of circulating miRNA characteristics as a noninvasive biomarker for the early detection of cancers has been reported. Ng et al examined the expression level of several serum miRNAs in patients with CRC and matched healthy controls. It was found that reduced serum miR-139-3p could serve as a biomarker for the early diagnosis of this disease with higher specificity and sensitivity.32 In addition, certain miRNAs have been identified as modulators of the tumor immune response. For example, miR-9 is frequently overexpressed in certain types of malignancies, and can contribute to the immunoescape of cancer cells via the downregulation of major histocompatibility complex class I.33 Meanwhile, clinical evidence has demonstrated that the expression profile of miRNAs varies significantly among normal tissues, cancer tissues, and different stages of cancers, and that some miRNAs can be considered promising biomarkers for the prediction of tumor behavior and survival time.34 Among these, miR-101 has long attracted numerous researchers’ attention, and relevant studies indicate that miR-101 frequently undergoes downregulation and may represent a tumor suppressor in various malignancies. In the last few years, decreased miR-101 expression has been connected to bad clinical outcomes in several tumor types, including NSCLC, HCC, bladder cancer, breast cancer, thyroid cancer,35 and oral cancer.36
As a tumor suppressor, miR-101 plays a crucial role in cancer biology via the targeting of various genes, which are shown in Table 4. In particular, many studies on diverse tumors have reported that EZH2, a mammalian histone methyltransferase, represents the main target gene of miR-101 and that decreased miR-101 with overexpression of EZH2 closely relates to cancer progression.37,38 However, miR-101 also targets other key genes involved in vital steps of tumorigenesis, such as ROCK1,39 CXCR7,40 MCL1,41 and RAC1.42 This targeting led to inhibited proliferation, migration, and invasion and promoted apoptosis for cancer cells. Additionally, reduced miR-101 expression contributing to the chemo- and radioresistance of cancer cells has been uncovered by researchers.43,44 Therefore, it is reasonable that miR-101 be regarded as a promising therapeutic target and a novel biomarker for tumor prognosis. As far as we know, a meta-analysis concerning the prognostic significance of decreased miR-101 on patients’ survival status has not been conducted. Nonetheless, the majority of articles have concluded that loss of miR-101 expression correlates with a poor outcome for cancer patients. However, there are still a few studies that have reported opposite results, suggesting that thus far, the prognostic value of miR-101 in various cancers is controversial and inconclusive. Therefore, we performed this comprehensive meta-analysis on the basis of gathering all related articles focusing on the associations between reduced miR-101 and cancer prognosis.
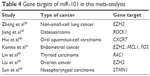  | Table 4 Gene targets of miR-101 in this meta-analysis |
In the meta-analysis, amounting to 12 studies with 2,088 cases, the impact of reduced miR-101 expression on OS in a series of malignancies yielded a pooled HR of 2.15 (95% CI 1.71–2.7), which suggested that decreased miR-101 was an unfavorable indicator and intimately related to deteriorated OS for patients suffering a variety of cancers. The existing heterogeneity regarding OS was quantified as 41.2% (I2), and for minimization of that, subgroup analysis was carried out on several factors that may have introduced potential heterogeneity to the combined results. When grouped by cancer types, decreased miR-101 predicted poorer survival status in NSCLC (HR 1.69, 95% CI 1.01–2.83) and HCC (HR 2.18, 95% CI 1.41–3.37), but not significantly in GB (HR 1.63, 95% CI 0.94–2.82). Similar synthesized results showed no obvious changes in heterogeneity, and were obtained from stratified analysis with respect to ethnicity, specimen source, and detection method, which further confirmed the prognostic value of miR-101 in those subgroups. However, compared to the overall pooled HR value, the associations of miR-101-expression loss were strongly accompanied with relatively smaller heterogeneity in the following subgroups: sample size >100, HR value reported, and multivariate analysis used. In addition, no obvious asymmetry observed in Begg funnel plots together with Egger’s test (P=0.987) indicated that no significant publication bias was present for OS. The result of sensitivity analysis for OS further confirmed the reliability of our results. To summarize, these results eventually revealed that decreased miR-101 was significantly associated with deteriorated OS in a number of carcinomas.
As similar definitions and a limited number of studies were included, survival indices, including DFS, RFS, and PFS, were combined to determine the predictive value of miR-101. In all four studies, 389 patients were recruited. Our results indicated that decreased miR-101 expression did not show a significant impact on DFS/RFS/PFS (HR 1.59, 95% CI 0.83–3.03), but the trend was positive. This insignificant result was most likely due to the limited number of studies included, and thus further research is warranted to identify the prognostic value of miR-101 in terms of DFS/RFS/PFS. Interestingly, reduced miR-101 correlated closely with certain clinicopathological features, including tumor-differentiation degree, T classification, and TNM stage, with pooled ORs of 2.17 (95% CI 1.14–4.13), 5.25 (95% CI 3.39–8.12), and 6.18 (95% CI 3.79–10.09), respectively. This suggested that decreased miR-101 might be linked to tumor progression significantly. However, further analyses were not performed, due to a limited number of studies. Furthermore, other relevant studies should be conducted to clarify the underlying significance of miR-101 for prognostication on DFS, RFS, and PFS.
The results from the meta-analysis performed here have several marked implications. First, decreased miR-101 expression may function as an effective tumor biomarker to reflect postoperative patient OS in malignant tumors. Although there were eight kinds of cancer included in the present analysis, the prognostic value of reduced miR-101 may extend to other kinds of tumors if there is higher between-study consistency and relatively acceptable heterogeneity. Second, decreased miR-101 expression correlated markedly with certain special clinicopathological parameters, such as tumor differentiation degree, tumor classification, and TNM stage, which unraveled the potential usage of miR-101 to predict the likelihood of tumor progression and refine the current risk-stratification system. Third, the lack of miR-101 was more useful as a prognostic indicator in the Chinese population, on account of the vast majority of articles being carried out in the China region. Fourth, sample size >100 was more suitable to determine the prognostic significance of miR-101 deficiency in cancer patients, as the stratified analysis regarding sample size demonstrated that the correlation between reduced miR-101 expression and OS was stronger together with minor heterogeneity (I2=22.2%). Fifth, multivariate analysis may be a better fit for the prediction of cancer prognosis. In theory, multivariate analyses, which are thought to rule out the impact of other confounding factors, such as age, tumor size, tumor grade, and tumor stage, are better and more accurate than univariate analyses. In addition, subgroup analysis according to analysis model confirmed this viewpoint.
Based on the evidence presented in this meta-analysis, loss of miR-101 expression was closely associated with patient prognosis and could be an unfavorable prognostic biomarker in a variety of solid tumors. Something that requires more attention are the limitations in these analyses. First, despite performing a thorough search for the identification of related articles, the number of included studies, in particular for DFS/RFS/PFS, was relatively small. This devalued our conclusions to some extent. Second, most eligible studies were conducted in Chinese populations, and the predictive value of miR-101 needs to be validated via further research in other Asian countries, as well as other regions with distinct ethnic backgrounds, such as Europe, the Americas, and Africa. Third, as there were only a limited number of included studies for each cancer type, the results of some carcinomas were statistically insignificant and thus less powerful. Fourth, the literature was restricted to English-language papers, which probably introduced a language bias. Fifth, the HRs of some studies were extrapolated based on Tierney et al’s method,24 which is less reliable than those directly provided in the original articles. Furthermore, the cutoff values in the study were not uniform, and may represent a source of heterogeneity. Finally, some publication bias was inevitable, as positive results are more easily accepted by journals than negative or null results. Given these limitations, our results should be considered cautiously.
In conclusion, the present meta-analysis of published articles demonstrated that loss of miR-101 expression was associated with deteriorated OS in a variety of cancer types, especially HCC and NSCLC. Decreased miR-101 was also an indicator for poorer clinicopathological features. Nonetheless, our results should be interpreted carefully, due to the aforementioned limitations. Our findings and the prognostic value of miR-101 in all kinds of malignancies should be further confirmed by large-scale standard investigations.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Nowsheen S, Aziz K, Panayiotidis MI, Georgakilas AG. Molecular markers for cancer prognosis and treatment: have we struck gold? Cancer Lett. 2012;327(1–2):142–152. | ||
Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20(8):460–469. | ||
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. | ||
Fang W, Fan Y, Fa Z, et al. MicroRNA-625 inhibits tumorigenicity by suppressing proliferation, migration and invasion in malignant melanoma. Oncotarget. 2017;8(8):13253–13263. | ||
Gu W, Zhan H, Zhou XY, et al. MicroRNA-22 regulates inflammation and angiogenesis via targeting VE-cadherin. FEBS Lett. 2017;591(3):513–526. | ||
Dong S, Yin H, Dong C, et al. Predictive value of plasma microRNA-216a/b in the diagnosis of esophageal squamous cell carcinoma. Dis Markers. 2016;2016:1857067. | ||
Ma YJ, Ha CF, Bai ZM, Li HN, Xiong Y, Jiang J. Overexpression of microRNA-205 predicts lymph node metastasis and indicates an unfavorable prognosis in endometrial cancer. Oncol Lett. 2016;12(6):4403–4410. | ||
Tang XR, Wen X, He QM, et al. MicroRNA-101 inhibits invasion and angiogenesis through targeting ITGA3 and its systemic delivery inhibits lung metastasis in nasopharyngeal carcinoma. Cell Death Dis. 2017;8(1):e2566. | ||
Lei Y, Li B, Tong S, et al. miR-101 suppresses vascular endothelial growth factor C that inhibits migration and invasion and enhances cisplatin chemosensitivity of bladder cancer cells. PLoS One. 2015;10(2):e0117809. | ||
Ye Z, Yin S, Su Z, et al. Downregulation of miR-101 contributes to epithelial-mesenchymal transition in cisplatin resistance of NSCLC cells by targeting ROCK2. Oncotarget. 2016;7(25):37524–37535. | ||
Luo L, Zhang T, Liu H, et al. MiR-101 and Mcl-1 in non-small cell lung cancer: expression profile and clinical significance. Med Oncol. 2012;29(3):1681–1686. | ||
Bao RF, Shu YJ, Hu YP, et al. miR-101 targeting ZFX suppresses tumor proliferation and metastasis by regulating the MAPK/Erk and Smad pathways in gallbladder carcinoma. Oncotarget. 2016;7(16):22339–22354. | ||
Li JT, Jia LT, Liu NN, et al. MiRNA-101 inhibits breast cancer growth and metastasis by targeting CX chemokine receptor 7. Oncotarget. 2015;6(31):30818–30830. | ||
Tian T, Mingyi M, Qiu X, Qiu Y. MicroRNA-101 reverses temozolomide resistance by inhibition of GSK3β in glioblastoma. Oncotarget. 2016;7(48):79584–79595. | ||
Li XP, Yuan ZB, Liu WJ, et al. CPEB1, a histone-modified hypomethylated gene, is regulated by miR-101 and involved in cell senescence in glioma. Cell Death Dis. 2013;4:e675. | ||
Zhang H, Qi F, Cao Y, Chen M, Zu X. Down-regulated microRNA-101 in bladder transitional cell carcinoma is associated with poor prognosis. Med Sci Monit. 2014;20:812–817. | ||
Li M, Tian L, Ren H, et al. MicroRNA-101 is a potential prognostic indicator of laryngeal squamous cell carcinoma and modulates CDK8. J Transl Med. 2015;13:271. | ||
Zheng F, Liao YJ, Cai MY, et al. Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PLoS Genet. 2015;11(2):e1004873. | ||
Zhang Y, Guo X, Xiong L, et al. MicroRNA-101 suppresses SOX9-dependent tumorigenicity and promotes favorable prognosis of human hepatocellular carcinoma. FEBS Lett. 2012;586(24):4362–4370. | ||
Slattery ML, Herrick JS, Pellatt DF, et al. Site-specific associations between miRNA expression and survival in colorectal cancer cases. Oncotarget. 2016;7(37):60193–60205. | ||
Lv X, Li J, Yang B. Clinical effects of miR-101 on prognosis of hepatocellular carcinoma and carcinogenic mechanism of anti-miR-101. Oncol Rep. 2016;36(4):2184–2192. | ||
Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ. 2009;339:b2535. | ||
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. | ||
Stang A. Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–605. | ||
Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. | ||
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. | ||
Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. | ||
Inamura K. Major tumor suppressor and oncogenic non-coding RNAs: clinical relevance in lung cancer. Cells. 2017;6(2):6020012. | ||
Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15(9):445. | ||
Inamura K, Ishikawa Y. MicroRNA in lung cancer: novel biomarkers and potential tools for treatment. J Clin Med. 2016;5(3):5030036. | ||
Ng L, Wan TM, Man JH, et al. Identification of serum miR-139-3p as a non-invasive biomarker for colorectal cancer. Oncotarget. 2017;8(16):27393–27400. | ||
Gao F, Zhao ZL, Zhao WT, et al. miR-9 modulates the expression of interferon-regulated genes and MHC class I molecules in human nasopharyngeal carcinoma cells. Biochem Biophys Res Commun. 2013;431(3):610–616. | ||
Zheng Q, Chen C, Guan H, Kang W, Yu C. Prognostic role of microRNAs in human gastrointestinal cancer: a systematic review and meta-analysis. Oncotarget. Epub 2017 Mar 29. | ||
Zhao H, Tang H, Huang Q, et al. MiR-101 targets USP22 to inhibit the tumorigenesis of papillary thyroid carcinoma. Am J Cancer Res. 2016;6(11):2575–2586. | ||
Wu B, Lei D, Wang L, et al. MiRNA-101 inhibits oral squamous-cell carcinoma growth and metastasis by targeting zinc finger E-box binding homeobox 1. Am J Cancer Res. 2016;6(6):1396–1407. | ||
Varambally S, Cao Q, Mani RS, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008;322(5908):1695–1699. | ||
Zhang JG, Guo JF, Liu DL, Liu Q, Wang JJ. MicroRNA-101 exerts tumor-suppressive functions in non-small cell lung cancer through directly targeting enhancer of Zeste homolog 2. J Thorac Oncol. 2011;6(4):671–678. | ||
Jiang R, Zhang C, Liu G, Gu R, Wu H. MicroRNA-101 inhibits proliferation, migration and invasion in osteosarcoma cells by targeting ROCK1. Am J Cancer Res. 2017;7(1):88–97. | ||
Hui Y, Li Y, Jing Y, Feng JQ, Ding Y. miRNA-101 acts as a tumor suppressor in oral squamous cell carcinoma by targeting CX chemokine receptor 7. Am J Transl Res. 2016;8(11):4902–4911. | ||
Konno Y, Dong P, Xiong Y, et al. MicroRNA-101 targets EZH2, MCL-1 and FOS to suppress proliferation, invasion and stem cell-like phenotype of aggressive endometrial cancer cells. Oncotarget. 2014;5(15):6049–6062. | ||
Lin X, Guan H, Li H, et al. miR-101 inhibits cell proliferation by targeting Rac1 in papillary thyroid carcinoma. Biomed Rep. 2014;2(1):122–126. | ||
Liu L, Guo J, Yu L, et al. miR-101 regulates expression of EZH2 and contributes to progression of and cisplatin resistance in epithelial ovarian cancer. Tumour Biol. 2014;35(12):12619–12626. | ||
Sun Q, Liu T, Zhang T, et al. MiR-101 sensitizes human nasopharyngeal carcinoma cells to radiation by targeting stathmin 1. Mol Med Rep. 2015;11(5):3330–3336. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



