Back to Journals » OncoTargets and Therapy » Volume 8
The prognostic significance of fibroblast growth factor receptor 4 in non-small-cell lung cancer
Authors Huang H, Feng H, Qiao H, Ren Z, Zhu G, liu X, Li P, guo S
Received 27 January 2015
Accepted for publication 11 March 2015
Published 22 May 2015 Volume 2015:8 Pages 1157—1164
DOI https://doi.org/10.2147/OTT.S81659
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Daniele Santini
Hong-ping Huang, Hui Feng, Hong-bo Qiao, Ze-xiang Ren, Ge-dong Zhu
Department of General Medicine, Linyi Hospital Affiliated to Shandong University, Linyi City, People’s Republic of China
Background: Fibroblast growth factor receptor 4 (FGFR4) has been proved to be correlated with progression and prognosis in many cancers. However, the significance of FGFR4 in non-small-cell lung cancer (NSCLC) is still not well elucidated.
Methods: In our experiment, we detected FGFR4 expression in 237 samples of NSCLC with immunohistochemistry, and further analyzed the correlation between FGFR4 and clinicopathologic features of NSCLC with chi-square test. Moreover, we evaluated the prognostic value of FGFR4 by Kaplan–Meier survival curve and Cox regression model. By regulating the expression of FGFR4 by overexpression or knockdown, we assessed the role of FGFR4 on NSCLC cell proliferation.
Results: FGFR4 expression was high in NSCLC (46.8%, 111/237). FGFR4 expression was significantly associated with tumor diameter (P=0.039). With univariate (P=0.009) and multivariate (P=0.002) analysis, FGFR4 was identified as an independent prognostic factor in NSCLC (P=0.009). Moreover, FGFR4 can promote the proliferation of NSCLC cell lines.
Conclusion: FGFR4 is an independent prognostic biomarker in NSCLC. FGFR4 can accelerate the proliferation of NSCLC cell lines, indicating FGFR4 could be a potential drug target of NSCLC.
Keywords: fibroblast growth factor 4, non-small-cell lung cancer, prognosis, proliferation
Introduction
Lung cancer is the most frequent cancer type, which leads to the most cases of cancer-related mortality in the world.1 The mortality and morbidity of lung cancer is still rising worldwide, especially in developing countries, partly because of the air pollution.2 Based on the clinical and histologic proof, lung cancer can be divided into small-cell lung cancer and non-small-cell lung cancer (NSCLC). NSCLC accounts for 85% of the primary bronchogenic carcinomas and has the highest mortality rate of malignant tumors in the world.3 NSCLC was traditionally considered as a single entity, with tumor stage regarded as the primary factor determining proper treatment. However, NSCLC consists of multiple and diverse histologic types and subtypes, with adenocarcinoma and squamous cell lung cancer. The molecular carcinogenesis of NSCLC features with multiple alterations of gene expression, which result in ectopic signaling pathways and biological behaviors.4–6 Although several biomarkers have been proved to be related with NSCLC angiogenesis, progression, or prognosis,7,8 there is still no effective predictive or prognostic biomarker clinically used, partly because most patients who are clinically diagnosed are in a middle or advanced stage. Consequently, the 5-year survival rate of NSCLC is as low as approximately 11%.9
Fibroblast growth factor receptors (FGFRs) consist of a family of four members, FGFR1, 2, 3, and 4, which have been demonstrated to be involved in the progression and prognosis of many kinds of cancers.10 They can activate downstream signaling pathways via binding with their ligands, which mostly are FGFs. Interestingly, FGFRs have diverse functions and downstream signaling pathways, which depend on binding different FGF ligands and alternative splicing. In the FGFR family, FGFR4 is distinguished because of its feature of high expression in liver.11 FGFR4 overexpression or mutation was demonstrated to be significantly associated with poor prognosis in many kinds of cancers, including hepatocellular carcinoma, prostate, breast, pancreatic, gynecologic gastric cancers, cholangiocarcinoma, and rhabdomyosarcomas.12–15 However, the significance of FGFR4 in lung cancer is still not well elucidated.
In our experiments, we detected the expression of FGFR4 in 237 cases of NSCLC with immunohistochemistry (IHC), and analyzed the correlation between FGFR4 and the clinicopathologic features in NSCLC. Furthermore, we identified the independent prognostic value of FGFR4 by univariate and multivariate analysis. After confirmation of FGFR4 as an independent prognostic factor, we performed cell function assays such as proliferation to evaluate the significance of FGFR4 in NSCLC progression.
Materials and methods
Patients and follow-up
Samples of NSCLC were obtained from the Pathological Department of Linyi Hospital and Yishui Central Hospital with prior approval of the patients and the Ethical Committees of these two hospitals. Three hundred and six patients were diagnosed with NSCLC and had undergone pulmonary lobectomy plus regional lymph node dissection from 2003 to 2007, and these patients made up the primary cohort. From the primary cohort, the validation cohort of 237 patients was selected with the following criteria: 1) available follow-up data; 2) available and enough samples; 3) R0 resection. Two senior pathologists confirmed the pathological diagnosis and selected proper sections for IHC. The paper was organized according to the REMARK instruction. Pathologic tumor-node-metastasis (pTNM) classification was based on the 7th International Union Against Cancer (2009).
IHC and evaluation
All NSCLC specimens were fixed by 10% formalin and embedded in paraffin, followed by cutting as serial sections deparaffinized with xylene. Slides were incubated in citrate buffer (pH =6.0) in a microwave oven for 30 minutes to achieve antigen retrieval. After that, specimens were incubated in 3% H2O2 in methanol for 20 minutes for endogenous peroxidase enzyme blocking. Primary antibody with dilution at 1:100 was used to incubate the tissue at 4°C overnight and then corresponding biotinylated secondary antibody and streptavidin–peroxidase complex was used to incubate at 37°C for 30 minutes. Finally, 3,3′-diaminobenzidine solution was used to make staining visible and hematoxylin was used for counterstaining. In the IHC test, the negative control was sampled with phosphate-buffered saline incubation instead of primary antibody, with other procedures all the same, while positive control was hepatocarcinoma sections, which had high FGFR4 expression.
The criterion of IHC evaluation was the combination of staining intensity and positive cell percentage. The percentage scores of immunoreactive cells were defined as follows: 0 for 10% positive cells; 1 for 10%–30% positive cells; 2 for 30%–50% cells; and 3 for 50% or more cells. The staining intensity can be described as follows: 0 for negative staining; 1 for weak staining; 2 for moderate staining; and 3 for strong staining. The final IHC score was defined as the multiplication of the staining intensity score and the positive cell score. The criteria for high FGFR4 expression and low FGFR4 expression were arbitrarily defined as ≥4 for high FGFR4 expression and <4 for low FGFR4 expression.
Cell culture and reagents
Two kinds of NSCLC adenocarcinoma cell lines (A549 and H1299) and two squamous carcinoma cell lines (SK-MES-1 and H520), as well as HepG2 and HEK293 cells, were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, People’s Republic of China). A549, H1299, and H520 cells were cultured in RPMI-1640, while SK-MES-1, HepG2, and HEK293 were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM), supplemented with 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin. RPMI-1640, DMEM, penicillin, streptomycin, fetal bovine serum, and trypsin were purchased from Thermo Fisher Scientific (Waltham, MA, USA). Anti-FGFR4 and anti-β-actin primary antibody were obtained from Santa Cruz Biotechnology Inc. (Dallas, TX, USA). 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) kit was from Sangon Company (Shanghai, People’s Republic of China).
Immunoblotting
Immunoblotting was used to detect the protein expression in NSCLC cells. Briefly, cells were lysed with RIPA lysis buffer on ice, then scraped and transferred into an Eppendorf tube. After centrifugation at 10,000× g at 4°C for 30 minutes, the supernatant was transferred into another tube and protein concentration was tested by Bradford method, followed by mixing with 2× loading buffer and boiling for 10 minutes for protein denaturation. After electrophoresis with 10 μg total protein, the protein was transferred to a nitrocellulose filter membrane (PALL Corporation) and subsequently blocked with 5% defatted milk. The nitrocellulose membrane was then incubated in primary antibody diluted in 5% bovine serum albumin at 4°C overnight. The nitrocellulose membrane was finally incubated in corresponding HRP-tagged secondary antibody, and then visualized by addition of ECL reagent (EMD Millipore, Billerica, MA, USA). The semi-quantitation of the immunoblotting result was achieved using the software ImageJ (National Institutes of Health [NIH], Bethesda, MD, USA).
FGFR4 knockdown, overexpression, and transfection
FGFR4 small interfering RNA (Thermo Fisher Scientific) was used for FGFR4 knockdown. The siRNA sequences were designed referring to a previous study.15 The sense sequence was 5′-GCCGACACAAGAACAUCAUTT-3′, and the antisense sequence was 5′-AUGAUGUUCUUGUGUCGGCTT-3′. As for the scrambled oligonucleotide RNA, the sense sequence was 5′-UUCUCCGAACGUGUCACGUTT-3′ and the anti-sense sequence was 5′-ACGUGACACGUUCGGAGAATT-3′.
Plasmid of pFLAG-CMV-2 vector or pFLAG-CMV-2-FGFR4 was purchased from Genephama Company (Shanghai, People’s Republic of China). Transfection of siRNA or plasmid was accomplished using Lipofectamine 2000 (Thermo Fisher Scientific), according to the transfection manual.
RNA extraction and real-time polymerase chain reaction analysis
Total RNA was purified from cancer tissue with TRIzol reagent after tissue grind and homogenation. Synthesis of cDNA was synthesized, and quantitative polymerase chain reaction (PCR) was realized by the StepOnePlus real-time PCR system (Thermo Fisher Scientific) using the SYBR Green method according to the manual. GAPDH (glyceraldehyde 3-phosphate dehydrogenase) was applied as an internal control. The sequences of primers used for real-time PCR experiments were designed following previous study and shown below:
- FGFR4 forward: 5′-CTGTGGCCGTCAAGATGCTCAA-3′
- FGFR4 reverse: 5′-ATGTTCTTGTGTCGGCCGATCA-3′
- GAPDH forward: 5′-GGGAAGGTGAAGGTCGGAGTC-3′
- GAPDH reverse: 5′-CCATGGGTGGAATCATATTGGAA-3′.
MTT assay
MTT assay was used to evaluate cell proliferation. Cells at logarithmic phase were trypsinized and passaged into 96-well plate at a density of 4,000 cells per well. After complete adhesion, cells were starved in serum-free medium and then incubated in normal medium with 10% fetal bovine serum, from which time was recorded. After incubation in normal medium for 0–72 hours, 10 μL MTT was added to the cells at concentration 10 mg/mL, followed by incubation at 37°C for 4–6 hours. The supernatant was discarded carefully and the crystals at the bottom were dissolved in 100 μL dimethyl sulfoxide. Absorbance at 490 nm was measured with a microplate reader (Molecular Devices LLC, Sunnyvale, CA, USA). Absorbance at 490 nm at the 0 time point was set as the baseline and the optical density at 490 nm of other groups was standardized with the ratio to baseline.
Statistical analysis
All statistical data were analyzed with SPSS 17.0 software (SPSS Inc., Chicago, IL, USA). The correlation between FGFR4 expression and the clinicopathologic features were analyzed with chi-square test. The univariate analysis was performed with the Kaplan–Meier survival curve method, and statistical differences were compared with a log-rank test. The multivariate analysis was carried out with Cox regression model. In experiments in vitro, the difference of the mean value between different groups was analyzed with Student’s t-test. P<0.05 was considered to be statistically significant.
Results
Patient characteristics and FGFR4 expression
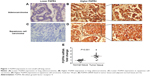
The validation cohort consisted of 237 NSCLC patients. The age of patients ranged from 33 to 78 years old, with median age 60 years (Table 1). Most patients (65%) were male in the validation cohort. The clinicopathologic parameters, including tumor size, lymph node metastasis, histological type, differentiation, and smoking, were according to hospital and surgical records. The cohort was divided into FGFR4 high-expression and low-expression groups according to the IHC criteria described in “Materials and methods”. In our study, FGFR4 expression was mainly observed in both cytoplasm and membrane (Figure 1A–D). Statistically, the rate of higher FGFR4 expression was 46.8% (111/237) in NSCLC. To compare the FGFR4 expression in tumor tissue or adjacent normal tissue, we detected the FGFR4 mRNA level from 12 pairs of frozen samples of NSCLC with quantitative PCR. It turned out that FGFR4 mRNA in tumor tissues was significantly higher than mRNA in normal tissues (Figure 1E).
  | Table 1 Characteristics of patients |
Correlation between FGFR4 and clinicopathologic factors in NSCLC
To screen suspicious factors correlated to FGFR4 expression, we analyzed FGFR4 association with clinicopathologic factors in NSCLC with the chi-square method (Table 2). Consequently, we found the FGFR4 higher-expression group had more cases of larger tumor diameter (P=0.039), indicating that FGFR4 may play important role in NSCLC cell proliferation. However, no other significant correlations between FGFR4 and other factors were observed in our experiments except the tumor diameter.
  | Table 2 Correlation between FGFR4 and clinicopathologic parameters |
Prognostic value of FGFR4 in NSCLC
To evaluate the prognostic value of FGFR4 in NSCLC, we first analyzed the correlation between FGFR4 expression and the 5-year overall survival rate with univariate analysis (Table 3). With the Kaplan–Meier method, we demonstrated that FGFR4 higher expression was correlated to poorer prognosis in NSCLC (P=0.009). In the FGFR4 higher-expression group, the 5-year overall survival rate was 17.4% with average survival time 39.8 months, while in the lower-expression group, the 5-year overall survival rate was 54.8% with average survival time 62.1 months (Figure 2A). In addition, stage of lymph node metastasis was also defined as a prognostic factor in NSCLC (P=0.003). Positive lymphatic invasion had a poorer prognosis than negative lymphatic invasion (42.7% vs 52.7% survival rate) (Figure 2B).
  | Table 3 Univariate analysis of non-small-cell lung cancer |
Moreover, we further performed multivariate analysis with Cox regression model to detect whether FGFR4 was an independent prognostic factor in NSCLC (Table 4). Almost all clinicopathologic features were collected into the model, including sex, age, tumor size, differentiation, lymph node metastasis, smoking, and FGFR4 expression. With multivariate analysis, we confirmed FGFR4 as an independent prognostic factor (P=0.002), with 95% confidence interval 1.35–3.71 and hazard ratio 2.24. Additionally, stage of lymph node invasion was also defined as an independent prognostic factor (P=0.001), with 95% confidence interval 1.38–3.79 and hazard ratio 2.23. No other clinicopathological parameter was identified as an independent prognostic factor in the Cox regression model in NSCLC.
FGFR4 promotes NSCLC cell line proliferation
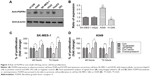
We demonstrated that FGFR4 expression was correlated with tumor diameter from analyzing clinical data with chi-square test, which indicated that FGFR4 may play an essential role in NSCLC proliferation. To confirm the result from clinical study with experiments in vitro, we performed MTT assay to evaluate the FGFR4 value in NSCLC cell line proliferation. We first detected FGFR4 expression in NSCLC cell lines to select proper cell models for FGFR4 regulation (Figure 3A). Four NSCLC cell lines were collected and used for FGFR4 detection, including adenocarcinoma cell lines A549 and H1299 and two squamous carcinomas cell lines, SK-MES-1 and H520, with HEK293 cells as negative control and HepG2 cells as positive control, which had high levels of FGFR4 expression. Among all the four cell lines, SK-MES-1 and H520 had similar FGFR4 expression, while A549 had the highest and H1299 had the lowest FGFR4 expression (Figure 3B). Moreover, we used MTT assay to access the regulation of FGFR4 in NSCLC proliferation. SK-MES-1 and A549 cells were transfected by scrambled RNA or siRNA to achieve FGFR4 knockdown and transfected by FLAG-FGFR4 or control p-FLAG-CMV2 to accomplish FGFR4 overexpression. After successful transfection, cells were starved overnight and then continued to be cultured in normal medium for 48 or 72 hours. The optical density of the control group at 490 nm was set as the baseline and optical density at 490 nm of other groups was measured by ratio with baseline. In both SK-MES-1 (Figure 3C) and A549 (Figure 3D), the overexpression of FGFR4 can enhance the proliferation, and the knockdown of FGFR4 can reduce the proliferation, which demonstrated that FGFR4 plays an important role in NSCLC and explains why the high-FGFR4 group had more cases of tumor diameter larger than 3 cm in the chi-square test.
Discussion
In our study, we systemically investigated the expression of FGFR4 in 237 cases of NSCLC for the first time and found that FGFR4 was associated with tumor diameter by chi-square test. With univariate and multivariate analysis, we identified FGFR4 as an independent prognostic factor of NSCLC. Furthermore, we proved that FGFR4 can promote NSCLC cell line proliferation, indicating that FGFR4 could be a potential molecular drug target of NSCLC chemotherapy.
In NSCLC, the FGFR family has been proved to be significantly associated with NSCLC progression. Amplification of FGFR1 can lead to poorer prognosis in early NSCLC,16 and several FGFR1 inhibitors have been demonstrated to suppress the growth of NSCLC cells overexpressing FGFR1 in NSCLC xenograft models.17,18 These inhibitors are mostly small-molecular inhibitors such as AZD4547, ponatinib (AP24534), and nintedanib (BIBF 1120). As a distinguishing feature, FGFR1/3 gene fusions are reported to define a molecular subset of NSCLC with distinct clinical characteristics.19 This kind of FGFR diversity and redundancy enormously increases the complexity and possibility of FGFR signaling pathway and the outcome it regulates.
FGFR4 is attracting more and more attention as a potential oncogene. FGFR4 gene amplification or protein overexpression is observed in several kinds of cancers. Different from FGFR1, the function and role of FGFR4 in NSCLC has not been well elucidated. Our study is the first research on FGFR4 effects on prognosis of NSCLC. We hope this study can trigger more interest in FGFR function and targeted therapy in NSCLC. Unfortunately, this research focused on clinical study of the correlation between FGFR4 and NSCLC prognosis, and did not determine the underlying molecular mechanism. More experiments are needed to elucidate downstream signaling pathways initiated by FGFR4 as well as the entire signaling network of FGFR4. In addition, the animal model is an essential tool for studying the role of biomarkers in cancer progression. We hope that our results in vitro can trigger further investigation of the role of FGFR4 in IHCC in vivo. Moreover, an important feature of FGFR4 is its polymorphism at the 388 site. Previous study indicates that the single nucleotide polymorphism of Arg388 is associated with prognosis in Japanese patients with NSCLC.20 Similar study of Arg388 allele function in the People’s Republic of China or other regions is also worthy of further experiments. The FGFR4 role in NSCLC in mouse models is not well known. Chemotherapy is an essential treatment for NSCLC, including preoperational chemotherapy and postoperational adjuvant chemotherapy. Currently used targeted chemical drugs are usually targeted at EGFR (gefitinib, erlotinib, or cetuximab) and VEGF (bevacizumab, ZD6474, or cediranib). Since the FGFR1 oncogenic role in NSCLC has been gradually revealed in recent years, chemical drugs targeting FGFR1 are more and more intriguing, and several FGFR1 inhibitors are in clinical trial. Our finding that FGFR4 is correlated to poor prognosis may bring new insight into NSCLC chemotherapeutic treatment.
Conclusion
We find that FGFR4 expression is significantly associated with tumor size and prognosis, and we identify FGFR4 as an independent prognostic factor in NSCLC. Moreover, FGFR4 can promote NSCLC cell proliferation in vitro. We hope our study can help with finding new chemotherapeutic drugs and increase the survival time and quality of life for patients with NSCLC.
Disclosure
The authors report no conflicts of interest in this work.
References
Jett JR, Cortese DA, Fontana RS. Lung cancer: current concepts and prospects. CA Cancer J Clin. 1983;33:74–86. | ||
Lu M, Tian H, Yue W, et al. TFIIB-related factor 2 over expression is a prognosis marker for early-stage non-small cell lung cancer correlated with tumor angiogenesis. PLoS One. 2014;9:e88032. | ||
Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med. 2008;359:1367–1380. | ||
Goldstraw P, Ball D, Jett JR, et al. Non-small-cell lung cancer. Lancet. 2011;378:1727–1740. | ||
Tang H, Tian H, Yue W, et al. Overexpression of LAPTM4B is correlated with tumor angiogenesis and poor prognosis in non-small cell lung cancer. Med Oncol. 2014;31:974. | ||
Li T, Kung HJ, Mack PC, Gandara DR. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J Clin Oncol. 2013;31:1039–1049. | ||
Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. | ||
Carlini MJ, Roitman P, Nuñez M, et al. Clinical relevance of galectin-1 expression in non-small cell lung cancer patients. Lung Cancer. 2014;84:73–78. | ||
Verdecchia A, Francisci S, Brenner H, et al; EUROCARE-4 Working Group. Recent cancer survival in Europe: a 2000–2002 period analysis of EUROCARE-4 data. Lancet Oncol. 2007;8:784–796. | ||
Kelleher FC, O’Sullivan H, Smyth E, McDermott R, Viterbo A. Fibroblast growth factor receptors, developmental corruption and malignant disease. Carcinogenesis. 2013;34:2198–2205. | ||
Song KH, Li T, Owsley E, Strom S, Chiang JY. Bile acids activate fibroblast growth factor 19 signaling in human hepatocytes to inhibit cholesterol 7alpha-hydroxylase gene expression. Hepatology. 2009;49: 297–305. | ||
Ye YW, Zhou Y, Yuan L, et al. Fibroblast growth factor receptor 4 regulates proliferation and antiapoptosis during gastric cancer progression. Cancer. 2011;117:5304–5313. | ||
Sugiyama N, Varjosalo M, Meller P, et al. Fibroblast growth factor receptor 4 regulates tumor invasion by coupling fibroblast growth factor signaling to extracellular matrix degradation. Cancer Res. 2010; 70:7851–7861. | ||
Taylor JG 6th, Cheuk AT, Tsang PS, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest. 2009;119:3395–3407. | ||
Xu YF, Yang XQ, Lu XF, et al. Fibroblast growth factor receptor 4 promotes progression and correlates to poor prognosis in cholangiocarcinoma. Biochem Biophys Res Commun. 2014;446:54–60. | ||
Cihoric N, Savic S, Schneider S, et al. Prognostic role of FGFR1 amplification in early-stage non-small cell lung cancer. Br J Cancer. 2014;110:2914–2922. | ||
Zhang J, Zhang L, Su X, et al. Translating the therapeutic potential of AZD4547 in FGFR1-amplified non-small cell lung cancer through the use of patient-derived tumor xenograft models. Clin Cancer Res. 2012;18:6658–6667. | ||
Ren M, Hong M, Liu G, et al. Novel FGFR inhibitor ponatinib suppresses the growth of non-small cell lung cancer cells overexpressing FGFR1. Oncol Rep. 2013;29:2181–2190. | ||
Wang R, Wang L, Li Y, et al. FGFR1/3 tyrosine kinase fusions define a unique molecular subtype of non-small cell lung cancer. Clin Cancer Res. 2014;20:4107–4114. | ||
Sasaki H, Okuda K, Kawano O, Yukiue H, Yano M, Fujii Y. Fibroblast growth factor receptor 4 mutation and polymorphism in Japanese lung cancer. Oncol Rep. 2008;20:1125–1130. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.