Back to Journals » OncoTargets and Therapy » Volume 13
The Potential Tumor Promotional Role of circVAPA in Retinoblastoma via Regulating miR-615-3p and SMARCE1
Authors Xu Q
Received 27 March 2020
Accepted for publication 4 July 2020
Published 5 August 2020 Volume 2020:13 Pages 7839—7849
DOI https://doi.org/10.2147/OTT.S254925
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sanjay Singh
This paper has been retracted.
Qibin Xu
Department of Ophthalmology, Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine (Hangzhou Red Cross Hospital), Hangzhou, Zhejiang Province, People’s Republic of China
Correspondence: Qibin Xu
Department of Ophthalmology, Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine (Hangzhou Red Cross Hospital), 208 Huancheng Road East, Hangzhou, Zhejiang Province 310003, People’s Republic of China
Tel +86 13989872710
Email [email protected]
Background: Growing evidence reveals that circular RNAs (circRNAs) play roles in cancer development. However, the effects and possible mechanisms of circRNAs in retinoblastoma (RB) are far from clear.
Methods: circVAPA expression pattern was identified by RT-qPCR. circVAPA induced effects on RB cells were tested by CCK-8, clone forming, flow cytometry and transwell assays. Bioinformatics assay, rescue experiments and dual-luciferase tests were applied for mechanism exploration. Additionally, mouse models were established for in vivo assays.
Results: circVAPA was upregulated in human RB specimen and RB cell lines, and was correlated with poor outcomes of Rb patients. Knockdown of circVAPA could suppress the malignant phenotypes of RB. Mechanistic experiments demonstrated that miR-615-3p could reverse the circVAPA induced effects on RB cells, and the downstream oncogene SMARCE1 was positively regulated by circVAPA via miR-615-3p. Further, in vivo analysis confirmed the findings.
Conclusion: In summary, circVAPA promoted RB proliferation and metastasis by sponging miR-615-3p, thereby upregulating SMARCE1. CircVAPA was a potential biomarker for Rb therapy.
Keywords: circular RNA, retinoblastoma, circVAPA, miR-615-3p, SMARCE1
Introduction
Retinoblastoma (RB) is the most common malignant tumor in children under five years old.1 RB is not sensitive to radiotherapy and chemotherapy. Although great efforts have been made to tackle this disease, the survival rate is under 5% in developing countries.2 Hence, exploring biological mechanisms of RB progression and finding out biomarkers and therapeutic approach are urgent for diagnose and therapy of this disease.
Circular RNAs (circRNAs) are a subclass of non-coding RNAs with single-strands, and attracted great attention recently.3,4 Most circRNAs are generated from exons with no protein-coding ability.5,6 CircRNAs were reported predominantly located in cytoplasm, where they sponge miRNAs, thereby releasing the target genes sequestered by miRNAs.7,8 Emerging evidence has reported the important roles of circRNAs in diverse cancer types.9 However, the function of circRNAs in RB has been rarely reported. circVAPA, derived from vesicle-associated membrane protein-associated protein A, was a novel circular RNA that was recently found associated with cancer progression. circVAPA was first found to play roles in colorectal cancer via interacting with miR-101.10 Liu et al observed that circVAPA was upregulated in hepatocellular cancer and enhanced cell proliferation.11 Zhou et al observed that circVAPA played as a tumor promoter via modulating miR-1615-5p in breast cancer development.12 Nevertheless, the role of circVAPA in other cancer types has not been reported, including in RB.
In our work, we observed the overexpression of circVAPA in RB tissues, as well as the oncogenic effects on RB cells by promoting cell proliferation, migration and invasion. Further, the underlying molecular mechanism was explored and we revealed that circVAPA acted as an oncogene in RB progression via sponging miR-615-3p, thereby positively modulating SMARCE1. This research may help provide novel targets for RB clinical diagnosis and therapy.
Methods
Clinical Samples
We obtained 50-paired RB tissue samples and adjacent normal ones from Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine. The involved patients received surgery between 2017 and 2019. We obtained written informed consent from every participant, and this study was approved by the Ethics Committee of Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine. Our study was conducted in accordance with the Declaration of Helsinki.
Cell Culture
Human retinal pigment epithelial cell line (ARPE-19) and human RB cell lines (WERI Rb1, hTERT-RPE1, SO-RB-50 and Y79) were provided by Cell Bank of Type Culture of Chinese Academy of Sciences (Shanghai, China). Cells were subjected to RPMI-1640 medium (Gibco, USA) with 10% FBS (Gibco, USA). Incubation was maintained at 37°C with 5%CO2.
Real-Time Quantitative PCR (RT-qPCR)
Total RNA was extracted by Trizol (Invitrogen, USA). In each sample, 2μg RNA was used to synthesize cDNA as the templates of RT-qPCR using MMLV (Promega, Beijing, China). RT-qPCR was carried out in triplicate using a PrimeScript RT reagent kit (Takara, Japan) on the pikoTM Thermal Cycler (ThermoFisher, USA). Relative expression levels of genes were calculated using the 2−ΔΔCt method. U6 and GAPDH were used as internal controls for miRNA and mRNA, respectively. The sequences of primers are presented in Table 1.
 |
Table 1 Primers of qRT-PCR |
Cell Transfection
siRNAs against circVAPA (siVAPA), miR-615-3p overexpression plasmids (miR-615-3p mimics), miR-615-3p inhibitor and corresponding negative controls were purchased from Integrated Biotech (Shanghai, China). Lipo3000 (Invitrogen, USA) was utilized for the subsequent transfection into cells.
Cell Viability Assay
CCK-8 (Sigma, USA) was utilized to measure cell viability. Each well of a 96-well plate was seeded with 2000 cells and followed by an incubation of described time. Cells were then added with CCK-8 solution and absorbance was measured at 450nm.
Colony-Forming Assay
Treated cells were seeded in 6-well plates with a density of 3000 cells per well, and cultured for 2 weeks. Then, methanol was utilized for fixing and 0.5% crystal violet was applied for staining. After 30 min, the number of colonies were counted.
Cell Apoptosis Assay
Cell apoptosis was detected using Annexin V/Cell apoptosis staining kit (LMAI Bio, Shanghai, China), according to the protocol. The cell apoptosis was tested using FACScan flow cytometer (BD Biosciences, San Jose, CA).
Transwell Assay
Transwell chamber (Corning, USA) was utilized for migration and invasion assays. Cells were seeded in the upper inserts filled with RPMI-1640, and the lower chambers were filled with complete medium (10% FBS). After 24 hours, cells suspended in the upper insets were removed. Cells in the lower chamber were fixed and stained with methanol and crystal violet. Migration cells were photographed using an inverted microscope (Olympus, Japan). For invasion assay, the upper chamber was precoated with Matrigel matrix (BD, USA).
Western Blot
Total protein was extracted from cells or tissues using RAPA lysis buffer (Thermo Fisher Scientific, USA). Equal amounts of each sample were loaded on 10% SDS PAGE gel and transferred on PVDF membranes. After blocked with 5% non-fat milk for an hour at room temperature, membranes were incubated at 4 °C overnight with specific primary antibodies against SMARCE1 (Abcam, 1:1000) and β-actin (Tiangen, 1:2000).
Immunohistochemical Staining
Tumors were treated with formalin and paraffin and sliced into 5 μM-thick sections. Xylene was utilized to deparaffinize the samples and ethanol was used for hydration. After blockage with serum for half an hour, samples were incubated with antibodies against SMARCE1 (1:200, Sigma) and Ki67 (1:500, Sigma) at 4°C overnight. Secondary antibodies were taken for another incubation at room temperature for an hour. DBA was utilized for color reaction.
Animal Model
Mice (8-week-old) were provided by Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine. Y29 cells carrying sh-circVAPA or sh-scramble (control group) were injected into flanks of nude mice. Tumor volumes were recorded weekly. Four weeks later, mice were sacrificed and tumors were removed for the subsequent assays. Animal study was approved by Ethics Committee of Zhejiang Hospital of Integrated Traditional Chinese and Western Medicine and conducted in accordance with the Guidelines for Animal Use in the National Institutes of Health.
Results
circVAPA Was Upregulated in RB and Indicated Poor Prognosis
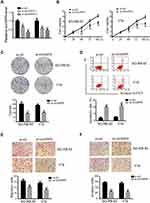
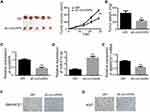
To identify expression patterns of circVAPA, qPCR was applied. As shown in Figure 1A and B, circVAPA expression levels were significantly increased both in RB patient tissue samples and cell lines. Correlations of circVAPA level with clinicopathological features were analyzed using Chi-squared test, and results (Table 2) indicated that circVAPA expression was correlated with tumor size (P = 0.032), FIGO stage (P = 0.025) and Lymph-node metastasis (P = 0.036). Moreover, higher circVAPA expression indicated poorer prognosis (Figure 1C). These findings suggested the potential oncogenic role of circVAPA.
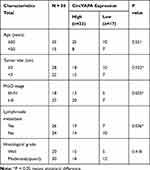 |
Table 2 Characteristics of RB Patients |
Knockdown of circVAPA Repressed Proliferation and Metastasis of RB Cells
To investigate the effects of circVAPA on RB, we used si-circVAPA for loss-of-function experiments. Figure 2A shows that both the two siRNAs against circVAPA exerted knockdown efficiencies. Functional experiments were performed following si-circVAPA (si-circVAPA-2) transfection. Cell viability was measured by CCK-8 assay, showing that the cell viabilities of SO-RB-50 and Y79 cells were decreased after transfection of si-circVAPA (Figure 2B). Colony-forming experiments also showed that the RB cell proliferation was suppressed with si-circVAPA transfection (Figure 2C). Whereas, the apoptotic rate of RB cells was increased upon circVAPA knockdown (Figure 2D). As metastasis was an evident feature of cancer cells, we further tested the effects of circVAPA on RB cell metastasis. As shown in Figure 2E and F, in circVAPA knockdown groups, the migration and invasion abilities were restrained, compared with those in control groups. All these results suggested that circVAPA might play as a tumor promotional role in RB progression.
circVAPA Served as a Sponge for miR-615-3p
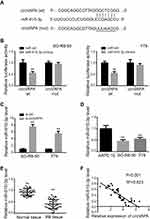
We next explored the possible molecular mechanism. We utilized CircBank to predict miR-615-3p as an interacting miRNA, with potential binding sites presented in Figure 3A. To confirm this prediction, we applied luciferase activity assay. As shown in Figure 3B, luciferase activity was suppressed with co-transfection of miR-615-3p and wild type circVAPA, but not with co-transfection of miR-615-3p and mutant circVAPA. Moreover, miR-615-3p expression was deceased when circVAPA was knocked down (Figure 3C). Further, we examined expressions of miR-615-3p in RB cell lines, observing the downregulated levels of miR-615-3p in SO-RB-50 and Y79 cell lines, in comparison with those in normal ARPE-19 cells (Figure 3D). Also, miR-615-3p was downregulated in RB tissues, in comparison with those in normal tissues (Figure 3E). Additionally, the expression levels of circVAPA and miR-615-3p in tumor tissues from 50 RB patients were detected by qPCR. Then, the correlation between circVAPA and miR-615-3p in RB tissues was determined using Pearson analysis. Results showed that miR-615-3p was negatively correlated with circVAPA (3F).
circVAPA Exerted Oncogenic Effects on RB Cells via Regulating miR-615-3p
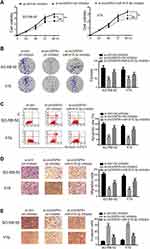
We further investigated whether miR-615-3p participated in circVAPA regulation on RB. si-circVAPA was co-transfected with/without miR-615-3p inhibitor, followed by functional experiments. We observed that miR-615-3p inhibitor attenuated the reduction of cell viability and colony number induced by si-circVAPA (Figure 4A and B). The aggravation on cell apoptosis induced by si-circVAPA was also mitigated by miR-615-3p inhibitor (Figure 4C). si-circVAPA downregulated the RB cell migration and invasion abilities, while miR-615-3p inhibitor could partially reverse these alterations (Figure 4D and E). These observations indicated that miR-615-3p was involved in the regulation.
circVAPA Upregulated SMARCE1 via miR-615-3p
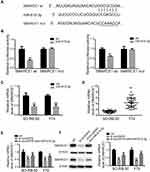
We utilized TargetScan network tool to predict SMARCE1 as the potential target of miR-615-3p (Figure 5A). We applied luciferase activity assay to confirm this prediction. As shown in Figure 5B, luciferase activity was suppressed with co-transfection of miR-615-3p and wild type 3ʹUTR of SMARCE1, but not with co-transfection of miR-615-3p and mutant 3ʹUTR of SMARCE1. Moreover, the expression level of SMARCE1 was downregulated by miR-615-3p overexpression (Figure 5C). These results confirmed that miR-615-3p directly interacted with SMARCE1. In addition, we observed that SMARCE1 was upregulated in RB tissues, in comparison with those in normal tissues (Figure 5D). Further, the mRNA and protein expression levels of SMRCE1 were downregulated by si-circVAPA, while miR-615-3p inhibitor could partially reverse these alterations (Figure 5E and F). All the above observations indicated that circVAPA sponged miR-615-3p to positively regulate SMARCE1 gene expression, which might be the underlying mechanism.
circVAPA Silencing Hampered RB Tumorigenesis in vivo
Tumor xenografts were generated in mice to perform in vivo experiments. Y79 cells expressing sh-circVAPA or sh-scramble (ctrl) were injected into mice. Tumor size and tumor weight were detected weekly and 4 weeks later, respectively (Figure 6A and B). After tumors were removed 4 weeks later, we examined the gene expressions in tumor tissues. RT-qPCR results showed that circVAPA and SMARCE1 were downregulated in circVAPA-depleted mice, while miR-615-3p was upregulated (Figure 6C–E). Immunohistochemistry assays were further performed to determine SMARCE1 and Ki67 expression levels. SMARCE1 was downregulated in sh-circVAPA mice, in line with the qPCR results we found (Figure 6F). Ki67, a cell proliferation marker, was also lowly expressed in sh-circVAPA mice (Figure 6G). The in vivo observations were consistent with those found in cell lines.
Discussion
RB is of high mortality rate that was commonly happened in children under five years old.13 Despite the great efforts in treating this disease, the survival rate is very low, especially in less developed countries.14 Hence, it is urgent to find out new approaches to overcome RB. Emerging evidence showed that circRNAs were dysregulated and played as tumor promotional or anti-cancer roles in cancer progressions.15 As to RB, there existed several reports demonstrating the relationship between circRNAs and RB development. For example, circ0001694 was revealed to overexpressed in human RB tissue and indicated poor survival rate by modulating AKT/mTOR signaling pathway.16 Circ0006168 was reported to activate S6K/S6 signaling and modulate miR-384/RBBP7, thereby contributing to RB cell viability and metastasis.17 Thus, in this study, we aimed to investigate the influence of circRNA in RB pathogenesis.
In this study, we applied qRT-PCR to determine the expression profile of circVAPA in RB. Results showed that circVAPA was highly expressed in RB tissue and cell lines. This result prompted us to explore the function of circVAPA in RB further. In vitro, knockdown of circVAPA inhibited RB cell viability and colony formation ability, as well as enhanced cell apoptosis. Moreover, the migration and invasion of SO-RB-50 and Y79 cells were significantly decreased after si-circVAPA were transfected. Of note, we detected the cell apoptosis, migration and invasion 24 h after transfection, when the cell viability had not been affected (the cell viability curve shown in Figure 2B). Thus, the reduced cell viability would not influence the phenotypic changes. These results suggested that circVAPA exerted a significant influence on RB cell proliferation, migration and invasion.
Further, we aimed to explore the possible mechanisms. More and more reports confirmed that circRNAs were rich in miRNA binding sites and likely to function by acting as sponges of miRNAs, thereby releasing the downstream target genes of miRNAs.18,19 For example, BCRC-3 inhibited bladder cancer through sequestering miR-182-5p and positively regulating p27.20 Circ103869 downregulated miR-532-3p and released FOXO4 mRNA, thereby contributing to colorectal cancer cell proliferation.21 Circ0084043 upregulated Snail via sponging miR-153-3p to promote malignant melanoma.22 In this study, we predicted the binding sites of circVAPA and miR-615-3p by informatics software and confirmed that circVAPA could bind to miR-615-3p by luciferase reporter activity. Moreover, we detected the expression pattern of miR-615-3p in RB cell lines and tissues, and found that miR-615-3p was lowly downregulated in RB cell lines and tissues, which was in line with the previous prediction. Also, in RB tissues, there existed a negative correlation between circVAPA and miR-615-3p analyzed by Pearson correlation analysis. Further, we conducted functional experiments to confirm that miR-615-3p was involved in the regulation mechanisms. si-circVAPA was co-transfected with miR-615-3p inhibitor or ctrl inhibitor. Then, CCK-8, colony formation assay, apoptosis assay and transwell assay were conducted. We found that si-circVAPA suppressed RB cell proliferation, migration and invasion, as well as promoted RB cell apoptosis, while miR-615-3p inhibitor could partially reverse these changes. These results suggested that circVAPA regulated RB cell proliferation and metastasis by sponging miR-615-3p.
MicroRNAs are known to bind to target genes’ 3ʹUTR, thus downregulate their expressions.23 Many miRNAs participate in cancer progression through repressing target mRNA expressions. For example, miR-376a-3p targeted KLF15 to promote colorectal cell proliferation and metastasis.24 miR-498 promoted RB cell proliferation and inhibited cell apoptosis via targeting CCPG1.25 Notch 1 and PAX6 were suppressed to express normally by miR-433, thereby hampering RB progression.26 Herein, we showed SMARCE1 as a direct target of miR-615-3p and found that SMARCE1 was highly expressed in RB tissue. SMARCE1 was a tumor promoter in many types of cancers, such as ovarian cancer,27 breast cancer,28 gastric cancer,29 and hepatoma carcinoma.30 In the present work, we predicted SMARCE1 as a target of miR-615-3p by TargetScan software, followed by luciferase reporter activity assays to confirm the prediction. In vivo, we detected the expression of SMARCE1, and found that SMARCE1 was upregulated in RB tissues, which might be inhibited by miR-615-3p. Furthermore, we wanted to investigate whether circVAPA could regulate SMARCE1 via miR-615-3p. si-circVAPA was co-transfected with miR-615-3p inhibitor or ctrl inhibitor, and SMARCE1 expression was detected. Results showed that SMARCE1 was suppressed by si-circVAPA, while the suppression was partially reversed by miR-615-3p inhibitor. These findings indicated that miR-615-3p/SMARCE1 axis might be involved in circVAPA regulated RB progression. To conform this supposition, we carried out in vivo experiments. Tumors in circVAPA knockdown group were smaller and lighter, in comparison with those in the control group. Importantly, miR-615-3p expression level was upregulated in circVAPA knockdown mice, while SMARCE1 expression level was downregulated. All the results suggested that circVAPA promoted RB progression via regulating miR-615-3p/SMARCE1.
Conclusion
CircVAPA promoted RB cell proliferation, migration and invasion, as well as inhibited cell apoptosis. In terms of mechanism, circVAPA could positively regulate SMACE1 via sponging miR-615-3p. This finding might provide new targets for clinical diagnosis and therapy of RB.
Disclosure
The author declares that they have no conflict of interest in this work.
References
1. Dimaras H, Kimani K, Dimba EA, et al. Retinoblastoma. Lancet. 2012;379(9824):1436–1446. doi:10.1016/S0140-6736(11)61137-9
2. Singh G, Daniels AB. Disparities in retinoblastoma presentation, treatment, and outcomes in developed and less-developed countries. Semin Ophthalmol. 2016;31(4):310–316. doi:10.3109/08820538.2016.1154177
3. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
4. Wu J, Qi X, Liu L, et al. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi:10.1016/j.omtn.2019.04.011
5. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
6. Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
7. Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi:10.1038/nrg.2016.114
8. Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. doi:10.1261/rna.043687.113
9. Hou L-D, Zhang J. Circular RNAs: an emerging type of RNA in cancer. Int J Immunopathol Pharmacol. 2017;30(1):1–6. doi:10.1177/0394632016686985
10. Li X-N, Wang Z-J, Ye C-X, et al. Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother. 2019;112:108611. doi:10.1016/j.biopha.2019.108611
11. Liu C, Zhong X, Li J, et al. Circular RNA circVAPA promotes cell proliferation in hepatocellular carcinoma. Hum Gene Ther Clin Dev. 2019;30(4):152–159. doi:10.1089/humc.2019.079
12. Zhou S-Y, Chen W, Yang S-J, et al. Circular RNA circVAPA regulates breast cancer cell migration and invasion via sponging miR-130a-5p. Epigenomics. 2020;12(4):303–317. doi:10.2217/epi-2019-0124
13. Benavente CA, Dyer MA. Genetics and epigenetics of human retinoblastoma. Annu Rev Pathol. 2015;10(1):547–562. doi:10.1146/annurev-pathol-012414-040259
14. Ortiz MV, Dunkel IJ. Retinoblastoma. J Child Neurol. 2016;31(2):227–236. doi:10.1177/0883073815587943
15. Vo JN, Cieslik M, Zhang Y, et al. The landscape of circular RNA in cancer. Cell. 2019;176(4):869–881 e13. doi:10.1016/j.cell.2018.12.021
16. Xing L, Zhang L, Feng Y, et al. Downregulation of circular RNA hsa_circ_0001649 indicates poor prognosis for retinoblastoma and regulates cell proliferation and apoptosis via AKT/mTOR signaling pathway. Biomed Pharmacother. 2018;105:326–333. doi:10.1016/j.biopha.2018.05.141
17. Xie Z-F, Li H-T, Xie S-H, et al. Circular RNA hsa_circ_0006168 contributes to cell proliferation, migration and invasion in esophageal cancer by regulating miR-384/RBBP7 axis via activation of S6K/S6 pathway. Eur Rev Med Pharmacol Sci. 2020;24(1):151–163. doi:10.26355/eurrev_202001_19906
18. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. doi:10.1016/j.jbiotec.2016.09.011
19. Zhang X, Zhu M, Yang R, et al. Identification and comparison of novel circular RNAs with associated co-expression and competing endogenous RNA networks in pulmonary tuberculosis. Oncotarget. 2017;8(69):113571–113582. doi:10.18632/oncotarget.22710
20. Xie F, Li Y, Wang M, et al. Circular RNA BCRC-3 suppresses bladder cancer proliferation through miR-182-5p/p27 axis. Mol Cancer. 2018;17(1):144. doi:10.1186/s12943-018-0892-z
21. Bian L, Zhi X, Ma L, et al. Hsa_circRNA_103809 regulated the cell proliferation and migration in colorectal cancer via miR-532–3p / FOXO4 axis. Biochem Biophys Res Commun. 2018;505(2):346–352. doi:10.1016/j.bbrc.2018.09.073
22. Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502(1):22–29. doi:10.1016/j.bbrc.2018.05.114
23. Mohr AM, Mott JL. Overview of microRNA biology. Semin Liver Dis. 2015;35(1):3–11. doi:10.1055/s-0034-1397344
24. Wang Y, Jiang F, Xiong Y, Cheng X, Qiu Z, Song R. LncRNA TTN-AS1 sponges miR-376a-3p to promote colorectal cancer progression via upregulating KLF15. Life Sci. 2019;244:116936.
25. Yang L, Wei N, Wang L, et al. miR-498 promotes cell proliferation and inhibits cell apoptosis in retinoblastoma by directly targeting CCPG1. Childs Nerv Syst’. 2018;34(3):417–422. doi:10.1007/s00381-017-3622-8
26. Li X, Yang L, Shuai T, et al. MiR-433 inhibits retinoblastoma malignancy by suppressing Notch1 and PAX6 expression. Biomed Pharmacother. 2016;82:247–255. doi:10.1016/j.biopha.2016.05.003
27. Giannakakis A, Karapetsas A, Dangaj D, et al. Overexpression of SMARCE1 is associated with CD8+ T-cell infiltration in early stage ovarian cancer. Int J Biochem Cell Biol. 2014;53:389–398. doi:10.1016/j.biocel.2014.05.031
28. Sethuraman A, Brown M, Seagroves TN, et al. SMARCE1 regulates metastatic potential of breast cancer cells through the HIF1A/PTK2 pathway. Breast Cancer Res. 2016;18(1):81. doi:10.1186/s13058-016-0738-9
29. Liu H, Zhao Y-R, Chen B, et al. High expression of SMARCE1 predicts poor prognosis and promotes cell growth and metastasis in gastric cancer. Cancer Manag Res. 2019;11:3493–3509. doi:10.2147/CMAR.S195137
30. Wu H-J, Zhuo Y, Zhou Y-C, et al. miR-29a promotes hepatitis B virus replication and expression by targeting SMARCE1 in hepatoma carcinoma. World J Gastroenterol. 2017;23(25):4569–4578. doi:10.3748/wjg.v23.i25.4569
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.