Back to Journals » OncoTargets and Therapy » Volume 13
The Perspective of Diagnostic and Prognostic Values of Lipoxygenases mRNA Expression in Colon Adenocarcinoma
Authors Ruan GT, Gong YZ , Zhu LC, Gao F , Liao XW , Wang XK, Zhu GZ , Liao C , Wang S, Yan L, Xie HL, Zhou X, Liu JQ, Shao MN , Gan JL
Received 29 February 2020
Accepted for publication 29 August 2020
Published 23 September 2020 Volume 2020:13 Pages 9389—9405
DOI https://doi.org/10.2147/OTT.S251965
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Geoffrey Pietersz
Guo-Tian Ruan1 ,* Yi-Zhen Gong1 ,* Li-Chen Zhu,2 Feng Gao,1 Xi-Wen Liao,3 Xiang-Kun Wang,3 Guang-Zhi Zhu,3 Cun Liao,1 Shuai Wang,1 Ling Yan,1 Hai-Lun Xie,1 Xin Zhou,3 Jun-Qi Liu,3 Meng-Nan Shao,4 Jia-Liang Gan1
1Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China; 2Department of Immunology, School of Preclinical Medicine, Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China; 3Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China; 4Life Sciences Institute, Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jia-Liang Gan
Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China
Email [email protected]
Background: This study was mainly to explore and study the potential application of lipoxygenases (ALOX) family genes in the diagnostic and prognostic values of colon adenocarcinoma (COAD).
Methods: Data sets related to the ALOX genes of COAD were obtained from The Cancer Genome Atlas and the University of California, Santa Cruz Xena browser. Then, the relevant biological information was downloaded from the public data platform. Finally, the bioinformatics technologies and clinical verification were employed to comprehensively analyze the potential values of ALOX genes.
Results: The Pearson correlation analysis indicated that there were correlations among ALOXE3, ALOX5, ALOX12, and ALOX12B. The diagnostic receiver operating characteristic (ROC) curves suggested that ALOXE3 and ALOX12 had significant diagnosis in COAD: ALOXE3; P< 0.001, area under curve (AUC) 95%CI:=0.818 (0.773– 0.862) and ALOX12; P< 0.001, AUC 95%CI=0.774 (0.682– 0.807). Besides, the verification study indicated that ALOX12 had a diagnostic value in COAD. Finally, our multivariate survival analysis and comprehensive prognosis of ALOX genes in COAD suggested that the ALOXE3 and ALOX12 were associated with COAD overall survival: ALOXE3; P=0.025, HR 95%CI=1.765 (1.074– 2.901), ALOX12; P=0.046, HR 95%CI=1.680 (1.009– 2.796), and the low expression of ALOXE3 and ALOX12 had a favorable prognosis of COAD (all P< 0.05); on the contrary, the high regulation of them increased the risk of death.
Conclusion: In our study, we observed that the mRNA expressions of ALOX genes were associated with the diagnosis and prognosis of COAD. The results of the diagnostic analysis suggested that ALOX12 might have a diagnosis value in COAD. Besides, our comprehensive prognosis analysis indicated that ALOXE3 combined ALOX12 might serve as potential prognosis biomarkers for COAD.
Keywords: mRNA, colon adenocarcinoma, diagnostic, prognostic, The Cancer Genome Atlas, lipoxygenases
Introduction
Colorectal cancer (CRC) is a common malignant neoplasm, which more than one million people worldwide suffer from every year increasing the burden of public health. In newest cancer statistics, CRC was the third most commonly occurring cancer in men (66,000 cases, accounting for 10% of the total), women ranked second (570,000 cases, accounting for 9.4% of the total), and CRC ranked third in terms of incidence but second in terms of mortality around the world in 2018.1 CRC was the leading cause of death and morbidity in developed countries.2 Interestingly, the incidence of CRC had risen sharply in Asian countries over the past 20 years, and historically, the incidence of CRC in Asian countries was relatively low.3 Colon adenocarcinoma (COAD) is the most common pathological type of CRC, the occurrence of COAD has undergone a multistage process from normal mucosa to adenoma to cancer.4 Although great progress had been made in the diagnosis and treatment (including surgery, targeted therapy, radiotherapy, and chemotherapy) of COAD, approximately half the patients relapsed or died within five years. Poor prognosis and the clinical outcome resulted from the inability to detect COAD at an early stage.5–7 Therefore, there is an urgent need to develop potential new diagnostic and prognostic biomarkers and effective therapeutic targets in COAD.
Lipoxygenases have been previously widely found in fungi, plants and animals.8–12 The human ALOX family genes (ALOXE3, ALOX5, ALOX12, ALOX12B, ALOX15 and ALOX15B)13,14 encode six differential isoforms, and the most polyenoic fatty acids including arachidonic acid (C20:Δ4, n-6), linolenic acid, linoleic acid (C18:Δ2, n-6), alpha- (C18:Δ3, n-3) and gamma- (C18:Δ3, n-6), docosahexaenoic acid (C22:Δ6, n-3) and eicosapentaenoic acid (C20:Δ5, n-3) can act as the substrates for different LOX-isoforms in most mammalian cells.14 Members of the ALOX family genes have highly conserved gene and protein structures. The mammalian ALOX genes have consisted of 14 or 15 exons, the exon/intron boundaries are highly conserved and they bind tightly together. Except for the ALOX5 gene mapped to chromosome 10, all other ALOX genes are located in a joint gene cluster on chromosome 17.13 Mammalian lipoxygenases ALOXs are non-heme iron-containing dioxygenases14,16 that play roles in lipid metabolism which converts the polyunsaturated fatty acids (PUFAs) including at least two isolated cis-double bonds to corresponding hydroperoxides.17–19 These intermediates in the cellular system could be rapidly degraded into alcohol or converted into bioactive lipid mediators, which include lipoxins,20 leukotrienes,21 resolvins,22 hepoxilins,23 and others stewing from arachidonic acid (AA), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA).15 The biological function of ALOXs has two alternative concepts: firstly, they can change the structural and functional parameters by oxidizing the complex lipids and even lipid-protein assemblies (biomembranes, lipoproteins). This can result in epidermal differentiation, atherogenesis and erythropoiesis.24–27 Secondly, they play important roles in gene expression, cell proliferation, and carcinogenesis, which work through forming lipid peroxides.28–31
However, there was little related research of ALOX family genes in COAD using bioinformatics. For the first time, our study downloaded RNA-sequencing (RNA-Seq) associated with the ALOX family genes from The Cancer Genome Atlas (TCGA) database and used relevant bioinformatics to study the potential diagnostic and prognostic value of COAD.
Materials and Methods
Public Database Resource Mining and Bioinformatics Analysis
We downloaded the expression information of ALOX family genes and corresponding clinical parameters from the public database The Cancer Genome Atlas (TCGA; https://tcga-data.nci.nih.gov/, obtained on January 30, 2019). We further collected corresponding clinical parameters associated with the TCGA database from the University of California, Santa Cruz Xena browser (UCSC Xena: http://xena.ucsc.edu/, accessed October 15, 2018). To investigate the biological processes, functional enrichment, and pathways of the ALOX family genes, we performed the online networking tool of Database for Annotation, Visualization, and Integrated Discovery (DAVID) (https://david.ncifcrf.gov/, version 6.8, obtained June 23, 2019).32,33 The online network analysis tool contained two parts: gene ontology (GO) enrichment functional analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. The GO term included three function analyses: biological process (BP), molecular function (MF) and cellular components (CC). An enrichment P-value <0.05 was of statistical significance. We also further explored the GO term enrichment analysis of ALOX family genes by using the BINGO analysis tool in Cytoscape version 3.6.1.34
Meanwhile, we investigated the Pearson correlations among ALOX family genes via the co-expression matrix which performed by R (version 3.5.1; www.r‑project.org).35 Then we explored the gene interactions using online tools of the Gene Multiple Association Network Integration Algorithm (GeneMANIA, http://www.genemania.org/, obtained June 27, 2019)36,37 and Search Tool for the Retrieval of Interacting Genes (STRING, https://string-db.org/, obtained June 27, 2019)38,39 at the levels of gene and protein.
Gene Differential Expression and Diagnostic ROC Curve Analysis
We explored the mRNA expression levels of ALOX family genes in COAD tumor tissues and adjacent normal tissues based on TCGA mRNA expression data and presented them in violin plots. Then we investigated the differential expression of ALOX family genes in colon tumor tissues and colon normal tissues based on 275 tumors and 379 normal samples matching TCGA normal and GTEx data, which conducted by Gene Expression Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/index.html; obtained June 28, 2019).40 Moreover, the distributable expressions of ALOX genes were conducted by the online analysis tool of Metabolic gEne Rapid Visualizer (MERAV; http://merav.wi.mit.edu/; obtained June 28, 2019).41 To further explore the potential diagnostic values of the ALOX genes, we performed the receiver operating characteristic (ROC) curves by GraphPad Prism software.
Verifying the Expression of ALOXE3 and ALOX12 of COAD in the First Affiliated Hospital of Guangxi Medical University Cohort
Collecting the COAD Patient’s Tissue Samples
We collected 16 pairs of patient’s tumor tissues with COAD and corresponding adjacent normal tissue samples from the Department of Colorectal and Anal Surgery, the First Affiliated Hospital of Guangxi Medical University from April to June 2018 (Nanning, China). All the patients were diagnosed were COAD.
Detection of the Expression of ALOXE3 and ALOX12 by RT-qPCR
We collected and stored the tissues in the RNA protection solution and stored it in a −80°C refrigerator, then lysed the tissue with TRIzol® reagent (Invitrogen) to extract total RNA. The RNA was reverse-transcribed into cDNA (20 uL) using a reverse transcription kit (Applied Biosystems by Thermo Fisher Scientific, USA), and finally, the cDNA was mixed with the FastStart Universal SYBR Green Master (ROX) kit (Roche) to perform qPCR which worked according to the guide of Applied Biosystems QuantsudioTM Real-Time PCR System (Q6) (Applied Biosystems by Thermo Fisher Scientific). The reaction conditions were: pre-heating at 95°C for 10 min; repeating 40 cycles at 95°C for 15 seconds, 60°C for one minute and 95°C for 30 seconds; in the end, denaturation at 95°C for 15 seconds, 60°C for one minute, 95°C for 30 seconds, 60°C for 15 seconds. The primers (designed and synthesized by Sangon Biotech (Shanghai) Co., Ltd, China) we used as followed: GAPDH (upstream: 5ʹ-TGGTCCCTGCTCCTCTAAC-3ʹ; downstream: 5ʹ-GGCTCAATGGCGTACTCTC-3ʹ), ALOXE3 (upstream: 5ʹ-CAAGGACTCTTGGTACTGTAGC-3ʹ; downstream: 5ʹ-TAGCCTTCAATCCACTGATAGC-3ʹ) and ALOX12 (upstream: 5ʹ-GATCCGAGGAGAGAAGCAATAC-3ʹ; downstream: 5ʹ-TGAGTGTTCAGCAAGTGATACT-3ʹ). The relative expression of ALOXE3 and ALOX12 we performed was used the method of 2‑ ∆∆ Cq.42
Survival Analysis
We integrated the patient’s clinical parameters and consistent gene expression levels, and the high- and low-expression groups were categorized by the gene expression level of the median cutoff value. Next we adjusted the relevant prognostic clinical parameters associated with COAD using a multivariate COX proportional hazards regression model. Then we obtained the prognostic ALOX genes of COAD. Finally, the joint effects survival analysis of prognostic ALOX genes has further investigated the association with the prognosis of COAD patients.
Comprehensive Prognosis Analysis of ALOX Genes
Based on the multivariate survival analysis, we performed a comprehensive analysis of prognostic ALOX genes. We conducted a risk-scoring model by multiplying the prognostic significance of ALOX genes expression levels by the contribution coefficient (β). The relevant model formula was as follows: risk score= expression of gene 1 × β of gene 1 + expression of gene 2 × β 2 of gene 2 + … + expression of gene n × β n of gene n.43,44 The establishment of a risk model divided the risk score into two groups, namely the high-risk group and low-risk group. The univariate survival analysis was carried out by high and low-risk scores, while the time-dependent survival of ROC was constructed by survivalROC package in the R platform. Besides, we also constructed a nomogram through the R platform to analyze the relevant prognostic contribution for COAD.
Statistical Analysis
The differential expression of ALOX genes in tumor tissues and adjacent normal tissues was analyzed by unpaired t-test. The log rank test was performed by univariate Kaplan–Meier survival analysis, and the 95%CIs and HRs were used to examine univariate and multivariate survival analyses. All P<0.05 were considered statistically different. All statistical analysis was generated by SPSS Statistics software version 20.0. Pearson correlation analysis, nomogram and survivalROC map were obtained through the R platform.
Results
Public Database Resource Mining
We downloaded clinical data of 456 patients with COAD from the TCGA public data platform, including sample information from 480 tumor tissues and 41 adjacent normal tissues. We also obtained consistent clinical parameters from the UCSC database, which consisted of 284 males and 262 females. Then we integrated two databases and got clinical parameters including overall survival (OS), recurrence-free survival (RFS), survival status, age, gender, carcino-embryonic antigen (CEA), KRAS, lymphatic invasion, and TNM stage. We excluded those patients with repeated information and survival time of 0. Finally, 438 tumor tissue samples and 41 adjacent tissue samples were obtained (shown in Table 1). In the RFS cohort study, all clinical parameters except age and tumor location were correlated with RFS (all P<0.05). Age, gender and tumor location did not show association with OS (P>0.05), but the others were significantly in connection with OS cohort study (all P<0.05).
 |
Table 1 Baseline Patient Characteristics in TCGA Cohort |
Bioinformatics Analysis
To acquire a more comprehensive and in-depth cognizance of the functions and characteristics of the ALOX family genes, we made use of bioinformatics to investigate potential biological functions of ALOX genes. We applied the online tool of DAVID to perform biological enrichment function of ALOX family genes, which GO terms analysis suggested that the ALOX genes might take part in the lipoxygenase pathway and complex lipid metabolic process, and the KEGG pathway analysis showed that they might be involved in arachidonic acid metabolism and metabolic pathway. (Figure S1) Then we used the BiNGO tool in Cytoscape to investigate the biological network of GO term analysis in ALOX genes, the results suggested a similar result with GO terms analysis (Figure S2).
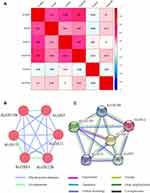
Meanwhile, we also explored the relationship between ALOX family genes through Pearson correlation analysis, and the results suggested that ALOXE3, ALOX5, ALOX12, and ALOX12B had relationships with each other (Figure 1A). When we explored the interaction network between ALOX family genes from the gene and protein level, we found that they had protein homology as well as co-expression at the gene and protein levels (Figure 1B and C).
Gene Differential Expression and Diagnostic ROC Curve Analysis
We investigated the differential expression of ALOX family genes based on TCGA, the results were shown in Figure 2. Meanwhile, we also downloaded the box plot of the ALOX genes from the GEPIA (Figure 3) and MERAV (Figure 4) online sites. In summary, the expression level of ALOXE3 in COAD tumor tissues was higher than that in adjacent normal tissues, but the expression of ALOX12 was relatively highly regulated in adjacent normal tissues. When we performed diagnostic ROC curve analyses, we found that ALOXE3; P<0.001, AUC, 95%CI=0.818 (0.773–0.862) and ALOX12; P<0.001, AUC, 95%CI=0.774 (0.682–0.807) had diagnostic values in COAD (Figure 5).
 |
Figure 2 Gene expression distribution of ALOX genes in TCGA. *P<0.05; **P<0.01; ***P<0.0001. |
 |
Figure 3 Expression levels of ALOX genes in tumor and normal tissues from the GEPIA website. (A), ALOXE3; (B), ALOX5; (C), ALOX12; (D), ALOX12B; (E), ALOX15; (F), ALOX15B. |
 |
Figure 4 Expression levels of ALOX genes in tumor and normal tissues from the MERAV website. (A), ALOXE3; (B), ALOX5; (C), ALOX12; (D), ALOX12B; (E), ALOX15; (F), ALOX15B. |
Verifying the Expression of ALOXE3 and ALOX12 of COAD in the First Affiliated Hospital of Guangxi Medical University Cohort
We performed and verified the expression of ALOXE3 and ALOX12 in COAD which showed that ALOXE3 was not significantly highly expressed in COAD tumor tissues but ALOX12 was significantly highly expressed in COAD adjacent tumor tissues. Meanwhile, the diagnostic ROC curves of ALOXE3 and ALOX12 in COAD suggested that the ALOX12 had a diagnostic value in COAD (P=0.029, AUC=0.713, 95%CI=0.544–0.881) but ALOXE3 had no diagnostic value in COAD (Figure 6).
Comprehensive Prognosis Survival Analysis
We adjusted the expression level of ALOX genes with prognostic clinical parameters, and the results suggested that ALOXE3; P=0.025, HR, 95%CI=1.765 (1.074–2.901) and ALOX12; P=0.046, HR, 95%CI=1.680 (1.009–2.796) had significant prognosis values in COAD OS (Table 2). Our survival curves analysis results suggested that the downregulation of ALOXE3 and ALOX12 both had a significantly good prognosis for COAD based on TCGA data (all P<0.05) (Figure 7).
 |
Table 2 Prognostic Values of ALOX Gene Expressions in COAD of TCGA Cohort |
 |
Figure 7 Kaplan–Meier survival curves for ALOX genes in COAD of the TCGA cohort. OS stratified by ALOXE3 (A); ALOX5 (B); ALOX12 (C); ALOX12B (D); ALOX15 (E); ALOX15B (F). |
Meanwhile, we performed a joint effects survival curve analysis by combining the expression of ALOXE3 and ALOX12 in the OS of COAD patients, the results suggested that both high expressions of ALOXE3 and ALOX12 increased the death risk in COAD (adjusted P=0.027), and on the contrary, both low expressions of ALOXE3 and ALOX12 decreased the death risk of COAD patients in TCGA cohort (adjusted P=0.009; adjusted HR=2.472; 95%CI=1.248–4.900) (Table 3, Figure 8).
 |
Table 3 Joint Effects Analysis of ALOXE3 and ALOX12 Expressions in COAD Patients OS |
 |
Figure 8 Joint effects analysis of ALOXE3 and ALOX12 of COAD OS in the TCGA cohort. |
We further explored the prognostic values of ALOXE3 and ALOX12. In our risk-scoring model, we categorized the risk score into two groups namely high-risk group and low-risk group, and then performed the survival status diagram and overall survival curve between the two groups, indicating that the low-risk group had fewer deaths and favorable prognoses. Otherwise, the five-year survival ROC curves performing by R platform suggested that all AUC >0.6 (shown in Figure 9). Our nomogram suggested that the TNM stage had the highest contribution to the prognosis of COAD, and we also observed that the other clinical parameters and prognosis genes of ALOXE3 and ALOX12 made a certain contribution of COAD survival prognosis in TCGA (Figure 10).
 |
Figure 10 Nomogram for predicting the prognosis of COAD patient. |
Discussion
COAD is one of the most serious diseases in the world that threaten human life, and it has a high incidence in Western countries and developed Asian countries.45,46 It is reported that genetic alterations had played a vital role in the tumorigenesis and development process of colon neoplasm.47,48 Lipoxygenases (ALOXs) are dioxygenases that catalyze the conversion of polyunsaturated fatty acids such as arachidonic acid to the corresponding peroxides. The ALOXs enzyme is expressed in epithelial cells, immune cells, and tumor cells, having various physiological and pathological functions including skin disorders, inflammation, and tumorigenesis.49 The metabolism of arachidonic acid which produced eicosanoids by lipoxygenase pathway played a significant role in tumor promotion, progression and metastasis.50,51
In our study, we first used bioinformatics analysis to explore the potential values of the ALOX family of genes. By analyzing the biological functions of ALOX family genes, our GO term and KEGG pathway analysis results indicated that ALOX genes might be involved in the process of the lipid metabolism process including lipoxygenase pathway and arachidonic acid metabolism. Moreover, the Pearson analysis showed that there were correlations among ALOXE3, ALOX5, ALOX12 and ALOX12B, the gene-gene and PPI networks showed that ALOXs had relationships between gene-gene and protein-protein expression levels.
Our differential expression analysis found that most of the genes (except for ALOX5 and ALOX15, all P<0.05) were significantly differentially expressed in COAD tumor and adjacent normal tissues, and the results also showed that the majority of genes were a low expression in tumor tissues, except for ALOXE3 gene. However, the outcome of diagnostic ROC curves, which were combined the TCGA cohort and Guangxi cohort, suggested that the ALOX12 had a significant diagnostic value in COAD, and the AUCs were above 0.7. As for the prognostic values of ALOX genes, our multivariate COX regression analysis suggested that the expression of ALOXE3 and ALOX12 had prognosis values associated with COAD OS. According to our comprehensives prognostic analyses, the results suggested that the low expression of ALOXE3 and ALOX12 had a favorable prognosis, which indicated that the high expression of them increased death risk. And our nomogram suggested that the ALOXE3 and ALOX12 made a certain contribution to the risk of COAD.
ALOXE3 is also known as ARCI3, E-LOX, eLOX3 and eLOX-3, which is a member of the lipoxygenase family. ALOXE3 is a hydroperoxide isomerase52 that synthesizes a unique type of epoxy alcohol (8R-hydroxy-11R, 12R-epoxyeicosa-5Z, 9E, 14Z-trienoic acid) from 12R-hydroperoxyeicosatetraenoic acid (12R-HPETE).53,54 Multiple database information (such as TCGA, GEPIA, and MERAV) has shown that ALOXE3 was highly expressed in tumor tissues. However, our verification cohort found that the ALOXE3 gene had no differential expression in COAD tumor tissues and adjacent normal tissues, this might be a bias caused by a small sample, which requires more experiments to verify. In addition, the ALOXE3 gene is a preferential expression in epithelial tissues, which plays a physiological role in the modulation or regulation of the proliferation and differentiation of epithelial cells and keratinocytes.55 It was reported that the mutation of ALOXE3 was associated with the development of autosomal recessive congenital ichthyosis (ARCI).53,54,56 But there was little known about ALOXE3 in the cancer field. We hypothesized that this gene might affect the transition from normal colonic epithelium to cancer.
ALOX12 (also known as LOG12, 12-LOX and 12S-LOX) utilizes arachidonic acid to synthesize 12(S)-hydroxyeicosatetraenoic acid [12(S)-HETE] which promotes tumorigenesis, transformation, and progression and metastasis.50,57,58 Many previous kinds of research had reported that the high expression of ALOX12 had been detected in various cancers, such as colorectal cancer,59,60 breast cancer,61,62 bladder cancer,63 prostate cancer,64,65 and others.58,66 Interestingly, our study and the related expression data obtained from the online tool of GEPIA of COAD found that the adjacent normal tissues had a high expression of ALOX12 when they were compared to the tumor tissues. Validation studies on ALOXE3 and ALOX12 by us indicated that ALOX12 was lowly expressed in COAD tumor tissues and had diagnostic value for COAD. Both cohort studies have consistent results. In Nigam et al’s research, they used (1-(14) C)-arachidonic acid to incubate uterine tissue homogenate in noncancer women and invasive cervical cancer patients under different conditions, and they analyzed the labeled reaction products by thin-layer chromatography and high-pressure liquid chromatography. They also used Western blot and immunohistochemical to investigate the expression level of 12-lipoxygenase protein. The results of Western blot suggested the highest concentrations and activities of 12-lipoxygenase were found in the exocervix, and immunohistochemical suggested the 12-lipoxygenase protein was unaltered while the bcl-2 gene was enhanced in invasive cervical carcinoma samples. Thus, they concluded that the process of carcinogenesis, the bcl-2 gene might mediate the ability of hydroperoxide reduction in uterine cervix tissue, and then metabolically inhibit 12-lipoxygenase activity.67 Moreover, a study by Gohara et al verified the expression of ALOX12, ALOX5, and ALOX15 in renal cell carcinoma (RCC) tumor tissues and normal tissues which found that the ALOX12 and ALOX5 genes were decreased and ALOX15 increased in tumor tissues. They found that this phenomenon was associated with the onset of cancer reversing with the progressing stage of the disease or the grade of tumor.68 Conversely, the ALOX12 might influence the growth and metastasis of cancer. In Klampfl et al’s study, they determined the expression of ALOX12 through quantitative RT-PCR from 50 colorectal tumor tissues, then they selected 34 tissue samples that had no inflammation, and the results found that no upregulation and even downregulation was observed in tumor samples. In addition, they found that upregulation of the ALOX12 in Caco2 and SW480 cells promoted the capacity of growth and metastasis in vivo.59 In Tan et al’s study, the genetic polymorphisms in 12-LOX 261Arg>Gln was determined by 1000 CRC patients and 1300 controls, which found that the increased risk of CRC was linked with the 12-LOX 261Arg>Gln/Gln genotype when it was compared to the Arg/Arg genotype (adjusted OR: 5 1.38, 95%CI=5 1.09–1.74).69 Colon cancer cell-derived 12-HETE induced contraction of cancer-associated fibroblasts by Ca2+ signaling, suggesting that 12-HETE might enhance CRC invasiveness.70 Similar results were also found in other cancers, such as acute myeloid leukemia (AML),71 prostate cancer,64 breast cancer72 and so on.
In our study, although we first used bioinformatics related methods to explore the potential diagnostic and prognostic values of the ALOX family genes in COAD, there were still some shortcomings. First, the related data were obtained from the public website, and the clinical parameters were not perfect. Second, the number of samples we had was relatively small and more samples are needed to further confirm. Third, the study still requires verified experiments at the tissue protein level, and we need more validation of this finding in vitro and in vivo. Fourth, the study needed a larger sample population and in a multicenter, multiregional, multiethnic population.
Conclusion
In this study, we used comprehensive bioinformatics methods and clinical verification to investigate the potential biological function and application of ALOX family genes in COAD, which suggested the mRNA expression levels of ALOX12 might be a diagnostic marker of COAD and ALOXE3 together with ALOX12 might serve as potential independent prognostic indicators for COAD. These results require necessary exploration and verification in the future.
Data Sharing Statement
The analyzed datasets generated during the study are available from The Cancer Genome Atlas (https://portal.gdc.cancer.gov/) and UCSC Xena (http://xena.ucsc.edu/).
Ethics Approval and Informed Consent
This study was based on the Declaration of Helsinki, all patients have signed informed consent forms, and our experimental protocol was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (No.2019 KY-E-001).
Acknowledgments
The authors would like to acknowledge the supported by the National Key Clinical Specialty Programs (General Surgery and Oncology) and the Key Laboratory of Early Prevention and Treatment for Regional High-Incidence-Tumor (Guangxi Medical University), Ministry of Education, China. The authors also thank the contributors of The Cancer Genome Atlas (https://cancergenome.nih.gov/) and UCSC Xena (http://xena.ucsc.edu/) for sharing the COAD dataset on open access.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Consent for Publication
Not applicable.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Pox CP. Controversies in colorectal cancer screening. Digestion. 2014;89(4):274–281. doi:10.1159/000363287
3. Pourhoseingholi MA. Increased burden of colorectal cancer in Asia. World J Gastrointest Oncol. 2012;4(4):68–70. doi:10.4251/wjgo.v4.i4.68
4. Bae JA, Kho DH, Sun EG, et al. Elevated coexpression of KITENIN and the ErbB4 CYT-2 isoform promotes the transition from colon adenoma to carcinoma following APC loss. Clin Cancer Res. 2016;22(5):1284–1294. doi:10.1158/1078-0432.CCR-15-0306
5. Aran V, Victorino AP, Thuler LC, Ferreira CG. Colorectal cancer: epidemiology, disease mechanisms and interventions to reduce onset and mortality. Clin Colorectal Cancer. 2016;15(3):195–203. doi:10.1016/j.clcc.2016.02.008
6. Yang Y, Li XJ, Li P, Guo XT. MicroRNA-145 regulates the proliferation, migration and invasion of human primary colon adenocarcinoma cells by targeting MAPK1. Int J Mol Med. 2018;42(6):3171–3180.
7. Hong J, Lu H, Meng X, Ryu JH, Hara Y, Yang CS. Stability, cellular uptake, biotransformation, and efflux of tea polyphenol (-)-epigallocatechin-3-gallate in HT-29 human colon adenocarcinoma cells. Cancer Res. 2002;62(24):7241–7246.
8. Grechkin A. Recent developments in biochemistry of the plant lipoxygenase pathway. Prog Lipid Res. 1998;37(5):317–352.
9. Gerwick WH. Structure and biosynthesis of marine algal oxylipins. Biochim Biophys Acta. 1994;1211(3):243–255. doi:10.1016/0005-2760(94)90147-3
10. De Petrocellis L, Di Marzo V. Aquatic invertebrates open up new perspectives in eicosanoid research: biosynthesis and bioactivity. Prostaglandins Leukot Essent Fatty Acids. 1994;51(4):215–229. doi:10.1016/0952-3278(94)90183-X
11. Funk CD. The molecular biology of mammalian lipoxygenases and the quest for eicosanoid functions using lipoxygenase-deficient mice. Biochim Biophys Acta. 1996;1304(1):65–84.
12. Yamamoto S, Suzuki H, Ueda N. Arachidonate 12-lipoxygenases. Prog Lipid Res. 1997;36(1):23–41. doi:10.1016/S0163-7827(97)00002-7
13. Funk CD, Chen XS, Johnson EN, Zhao L. Lipoxygenase genes and their targeted disruption. Prostaglandins Other Lipid Mediat. 2002;6869:303–312. doi:10.1016/S0090-6980(02)00036-9
14. Ivanov I, Kuhn H, Heydeck D. Structural and functional biology of arachidonic acid 15-lipoxygenase-1 (ALOX15). Gene. 2015;573(1):1–32. doi:10.1016/j.gene.2015.07.073
15. Haeggstrom JZ, Funk CD. Lipoxygenase and leukotriene pathways: biochemistry, biology, and roles in disease. Chem Rev. 2011;111(10):5866–5898. doi:10.1021/cr200246d
16. Brash AR. Lipoxygenases: occurrence, functions, catalysis, and acquisition of substrate. J Biol Chem. 1999;274(34):23679–23682. doi:10.1074/jbc.274.34.23679
17. Yamamoto S. Mammalian lipoxygenases: molecular structures and functions. Biochim Biophys Acta. 1992;1128(23):117–131. doi:10.1016/0005-2760(92)90297-9
18. Orafaie A, Matin MM, Sadeghian H. The importance of 15-lipoxygenase inhibitors in cancer treatment. Cancer Metastasis Rev. 2018;37(23):397–408.
19. Kuhn H, Humeniuk L, Kozlov N, Roigas S, Adel S, Heydeck D. The evolutionary hypothesis of reaction specificity of mammalian ALOX15 orthologs. Prog Lipid Res. 2018;72:55–74. doi:10.1016/j.plipres.2018.09.002
20. Hu S, Mao-Ying QL, Wang J, et al. Lipoxins and aspirin-triggered lipoxin alleviate bone cancer pain in association with suppressing expression of spinal proinflammatory cytokines. J Neuroinflammation. 2012;9:278. doi:10.1186/1742-2094-9-278
21. Savari S, Vinnakota K, Zhang Y, Sjolander A. Cysteinyl leukotrienes and their receptors: bridging inflammation and colorectal cancer. World J Gastroenterol. 2014;20(4):968–977.
22. Yoo S, Lim JY, Hwang SW. Resolvins: endogenously-generated potent painkilling substances and their therapeutic perspectives. Curr Neuropharmacol. 2013;11(6):664–676. doi:10.2174/1570159X11311060009
23. Brash AR, Yu Z, Boeglin WE, Schneider C. The hepoxilin connection in the epidermis. FEBS J. 2007;274(14):3494–3502. doi:10.1111/j.1742-4658.2007.05909.x
24. Krieg P, Rosenberger S, de Juanes S, et al. Aloxe3 knockout mice reveal a function of epidermal lipoxygenase-3 as hepoxilin synthase and its pivotal role in barrier formation. J Invest Dermatol. 2013;133(1):172–180. doi:10.1038/jid.2012.250
25. Epp N, Furstenberger G, Muller K, et al. 12R-lipoxygenase deficiency disrupts epidermal barrier function. J Cell Biol. 2007;177(1):173–182.
26. Schewe T, Wiesner R, Rapoport SM. Lipoxygenase from rabbit reticulocytes. Methods Enzymol. 1981;71(Pt):
27. Zhao L, Funk CD. Lipoxygenase pathways in atherogenesis. Trends Cardiovasc Med. 2004;14(5):191–195. doi:10.1016/j.tcm.2004.04.003
28. Wen Z, Liu H, Li M, et al. Increased metabolites of 5-lipoxygenase from hypoxic ovarian cancer cells promote tumor-associated macrophage infiltration. Oncogene. 2015;34(10):1241–1252. doi:10.1038/onc.2014.85
29. Cathcart MC, Lysaght J, Pidgeon GP. Eicosanoid signalling pathways in the development and progression of colorectal cancer: novel approaches for prevention/intervention. Cancer Metastasis Rev. 2011;30(34):363–385.
30. Nie D, Hillman GG, Geddes T, et al. Platelet-type 12-lipoxygenase in a human prostate carcinoma stimulates angiogenesis and tumor growth. Cancer Res. 1998;58(18):4047–4051.
31. Kelavkar UP, Nixon JB, Cohen C, Dillehay D, Eling TE, Badr KF. Overexpression of 15-lipoxygenase-1 in PC-3 human prostate cancer cells increases tumorigenesis. Carcinogenesis. 2001;22(11):1765–1773. doi:10.1093/carcin/22.11.1765
32. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi:10.1093/nar/gkn923
33. Dennis G
34. Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi:10.1093/bioinformatics/bti551
35. Wang X, Liao X, Yang C, et al. Identification of prognostic biomarkers for patients with hepatocellular carcinoma after hepatectomy. Oncol Rep. 2019;41(3):1586–1602.
36. Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi:10.1186/gb-2008-9-s1-s4
37. Franz M, Rodriguez H, Lopes C, et al. GeneMANIA update 2018. Nucleic Acids Res. 2018;46(W1):W60W64. doi:10.1093/nar/gky311
38. von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261. doi:10.1093/nar/gkg034
39. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362D368. doi:10.1093/nar/gkw937
40. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98w102. doi:10.1093/nar/gkx247
41. Shaul YD, Yuan B, Thiru P, et al. MERAV: a tool for comparing gene expression across human tissues and cell types. Nucleic Acids Res. 2016;44(D1):D560566.
42. Ruan GT, Gong YZ, Liao XW, et al. Diagnostic and prognostic values of CXC motif chemokine ligand 3 in patients with colon cancer. Oncol Rep. 2019. doi:10.3892/or.2019.7326
43. Liao X, Zhu G, Huang R, et al. Identification of potential prognostic microRNA biomarkers for predicting survival in patients with hepatocellular carcinoma. Cancer Manag Res. 2018;10:787–803. doi:10.2147/CMAR.S161334
44. Huang R, Liao X, Li Q. Identification and validation of potential prognostic gene biomarkers for predicting survival in patients with acute myeloid leukemia. Onco Targets Ther. 2017;10:5243–5254. doi:10.2147/OTT.S147717
45. Jiang H, Du J, Gu J, Jin L, Pu Y, Fei B. A 65gene signature for prognostic prediction in colon adenocarcinoma. Int J Mol Med. 2018;41(4):2021–2027.
46. Wang JY, Wang CL, Wang XM, Liu FJ. Comprehensive analysis of microRNA/mRNA signature in colon adenocarcinoma. Eur Rev Med Pharmacol Sci. 2017;21(9):2114–2129.
47. Mullany LE, Herrick JS, Wolff RK, Slattery ML. MicroRNA Seed Region Length Impact on Target Messenger RNA Expression and Survival in Colorectal Cancer. PLoS One. 2016;11(4):e0154177. doi:10.1371/journal.pone.0154177
48. Xing Y, Zhao Z, Zhu Y, Zhao L, Zhu A, Piao D. Comprehensive analysis of differential expression profiles of mRNAs and lncRNAs and identification of a 14-lncRNA prognostic signature for patients with colon adenocarcinoma. Oncol Rep. 2018;39(5):2365–2375.
49. Mashima R, Okuyama T. The role of lipoxygenases in pathophysiology; new insights and future perspectives. Redox Biol. 2015;6:297–310. doi:10.1016/j.redox.2015.08.006
50. Avis I, Hong SH, Martinez A, et al. Five-lipoxygenase inhibitors can mediate apoptosis in human breast cancer cell lines through complex eicosanoid interactions. FASEB J. 2001;15(11):2007–2009. doi:10.1096/fj.00-0866fje
51. Wang Y, Wang W, Sanidad KZ, Shih PA, Zhao X, Zhang G. Eicosanoid signaling in carcinogenesis of colorectal cancer. Cancer Metastasis Rev. 2018;37(23):257–267.
52. Yu Z, Schneider C, Boeglin WE, Marnett LJ, Brash AR. The lipoxygenase gene ALOXE3 implicated in skin differentiation encodes a hydroperoxide isomerase. Proc Natl Acad Sci U S A. 2003;100(16):9162–9167. doi:10.1073/pnas.1633612100
53. Jobard F, Lefevre C, Karaduman A, et al. Lipoxygenase-3 (ALOXE3) and 12(R)-lipoxygenase (ALOX12B) are mutated in non-bullous congenital ichthyosiform erythroderma (NCIE) linked to chromosome 17p13.1. Hum Mol Genet. 2002;11(1):107–113. doi:10.1093/hmg/11.1.107
54. Eckl KM, Krieg P, Kuster W, et al. Mutation spectrum and functional analysis of epidermis-type lipoxygenases in patients with autosomal recessive congenital ichthyosis. Hum Mutat. 2005;26(4):351–361. doi:10.1002/humu.20236
55. Krieg P, Marks F, Furstenberger G. A gene cluster encoding human epidermis-type lipoxygenases at chromosome 17p13.1: cloning, physical mapping, and expression. Genomics. 2001;73(3):323–330. doi:10.1006/geno.2001.6519
56. Eckl KM, de Juanes S, Kurtenbach J, et al. Molecular analysis of 250 patients with autosomal recessive congenital ichthyosis: evidence for mutation hotspots in ALOXE3 and allelic heterogeneity in ALOX12B. J Invest Dermatol. 2009;129(6):1421–1428. doi:10.1038/jid.2008.409
57. Huang Z, Xia L, Zhou X, Wei C, Mo Q. ALOX12 inhibition sensitizes breast cancer to chemotherapy via AMPK activation and inhibition of lipid synthesis. Biochem Biophys Res Commun. 2019;514(1):24–30.
58. Winer I, Normolle DP, Shureiqi I, et al. Expression of 12-lipoxygenase as a biomarker for melanoma carcinogenesis. Melanoma Res. 2002;12(5):429–434. doi:10.1097/00008390-200209000-00003
59. Klampfl T, Bogner E, Bednar W, et al. Up-regulation of 12(S)-lipoxygenase induces a migratory phenotype in colorectal cancer cells. Exp Cell Res. 2012;318(6):768–778. doi:10.1016/j.yexcr.2011.12.017
60. Li S, Zhao X, Wu Z, et al. Polymorphisms in arachidonic acid metabolism-related genes and the risk and prognosis of colorectal cancer. Fam Cancer. 2013;12(4):755–765. doi:10.1007/s10689-013-9659-2
61. Jiang WG, Douglas-Jones A, Mansel RE. Levels of expression of lipoxygenases and cyclooxygenase-2 in human breast cancer. Prostaglandins Leukot Essent Fatty Acids. 2003;69(4):275–281. doi:10.1016/S0952-3278(03)00110-8
62. Singh AK, Kant S, Parshad R, Banerjee N, Dey S. Evaluation of human LOX-12 as a serum marker for breast cancer. Biochem Biophys Res Commun. 2011;414(2):304–308. doi:10.1016/j.bbrc.2011.09.044
63. Yoshimura R, Matsuyama M, Tsuchida K, Kawahito Y, Sano H, Nakatani T. Expression of lipoxygenase in human bladder carcinoma and growth inhibition by its inhibitors. J Urol. 2003;170(5):1994–1999. doi:10.1097/01.ju.0000080296.54262.c8
64. Matsuyama M, Yoshimura R, Mitsuhashi M, et al. Expression of lipoxygenase in human prostate cancer and growth reduction by its inhibitors. Int J Oncol. 2004;24(4):821–827.
65. Nie D, Nemeth J, Qiao Y, et al. Increased metastatic potential in human prostate carcinoma cells by overexpression of arachidonate 12-lipoxygenase. Clin Exp Metastasis. 2003;20(7):657–663. doi:10.1023/A:1027302408187
66. De Armas R, Durand K, Guillaudeau A, et al. mRNA levels of enzymes and receptors implicated in arachidonic acid metabolism in gliomas. Clin Biochem. 2010;43(1011):827–835. doi:10.1016/j.clinbiochem.2010.03.017
67. Nigam S, Kumar GS, Sutherland M, et al. Metabolic suppression of platelet-type 12-lipoxygenase in human uterine cervix with invasive carcinoma. Int J Cancer. 1999;82(6):827–831. doi:10.1002/(SICI)1097-0215(19990909)82:6<827::AID-IJC10>3.0.CO;2-Q
68. Gohara A, Eltaki N, Sabry D, et al. Human 5-, 12- and 15-lipoxygenase-1 coexist in kidney but show opposite trends and their balance changes in cancer. Oncol Rep. 2012;28(4):1275–1282. doi:10.3892/or.2012.1924
69. Tan W, Wu J, Zhang X, et al. Associations of functional polymorphisms in cyclooxygenase-2 and platelet 12-lipoxygenase with risk of occurrence and advanced disease status of colorectal cancer. Carcinogenesis. 2007;28(6):1197–1201. doi:10.1093/carcin/bgl242
70. Stadler S, Nguyen CH, Schachner H, et al. Colon cancer cell-derived 12(S)-HETE induces the retraction of cancer-associated fibroblast via MLC2, RHO/ROCK and Ca(2+) signalling. Cell Mol Life Sci. 2017;74(10):1907–1921. doi:10.1007/s00018-016-2441-5
71. Ohgami RS, Ma L, Ren L, et al. DNA methylation analysis of ALOX12 and GSTM1 in acute myeloid leukaemia identifies prognostically significant groups. Br J Haematol. 2012;159(2):182–190. doi:10.1111/bjh.12029
72. Singh AK, Singh R, Naz F, et al. Structure based design and synthesis of peptide inhibitor of human LOX-12: in vitro and in vivo analysis of a novel therapeutic agent for breast cancer. PLoS One. 2012;7(2):e32521. doi:10.1371/journal.pone.0032521
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.