Back to Journals » Cancer Management and Research » Volume 11
The oncoprotein HBXIP facilitates metastasis of hepatocellular carcinoma cells by activation of MMP15 expression
Authors Zheng S, Wu H, Wang F , Lv J, Lu J, Fang Q, Wang F, Lu Y, Zhang S , Xu Y, Bao Q, Xie C , Yin Z
Received 19 December 2018
Accepted for publication 21 March 2019
Published 16 May 2019 Volume 2019:11 Pages 4529—4540
DOI https://doi.org/10.2147/CMAR.S198783
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Kenan Onel
Sen Zheng,1,* Huita Wu,2,* Fei Wang,1,* Jie Lv,1 Jing Lu,1 Qinliang Fang,1 Fuqiang Wang,1 Yuyan Lu,1 Sheng Zhang,1 Yaping Xu,3 Qing Bao,1 Chengrong Xie,1 Zhenyu Yin1
1Department of Hepatobiliary Surgery, Zhongshan Hospital, Xiamen University, Fujian Provincial Key Laboratory of Chronic Liver Disease and Hepatocellular Carcinoma, Xiamen 361004, Fujian, People’s Republic of China; 2Department of Oncology, Zhongshan Hospital, Xiamen University, Xiamen 361004, Fujian, People’s Republic of China; 3Key laboratory of functional and clinical translational medicine, Xiamen Medical College, Xiamen 361004, Fujian, People’s Republic of China
*These authors contributed equally to this work
Background: Due to the high recurrence and metastasis rate, the clinical outcomes of patients with hepatocellular carcinoma (HCC) are still unsatisfactory. Hepatitis B virus X-interacting protein (HBXIP) has been reported to play crucial roles in carcinogenesis.
Purpose: We aimed to reveal the functional significance and underlying mechanism of HBXIP in HCC metastasis.Methods: Cell transwell assay, in vivo metastasis model, real-time PCR, western blot analysis, luciferase reporter and chromatin immunoprecipitation assays were applied.
Results: Here, we detected the HBXIP expression level and determined its clinical significance in HCC. We found that HBXIP was significantly upregulated in HCC tissues, and correlated with vascular invasion, tumor metastasis and worse prognosis of HCC patients. HBXIP enhanced cell migration and invasion in vitro, and promoted the metastasis of HCC in vivo. Furthermore, we confirmed that HBXIP increased MMP15 expression through association with proto-oncogene c-myc. Depletion of c-myc abolished HBXIP-mediated MMP-15 upregulation. We also observed a positive correlation between HBXIP and MMP15 expression in HCC tissues.
Conclusion: Our results establish a novel function for HBXIP-MMP15 regulation in HCC metastasis and suggest its candidacy as a new prognostic biomarker and therapeutic target for HCC metastasis.
Keywords: c-myc, MMP15, HBXIP, metastasis
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent malignancies worldwide.1 The major risk factors for HCC include chronic infection with hepatitis B or C, alcoholic liver injury, exposure to dietary aflatoxin B and steatohepatitis.2 Despite the advancement of HCC therapy, such as surgical resection, chemotherapy, radiotherapy and liver transplantation, the clinical outcomes of patients with HCC remain unsatisfactory due to the high recurrence and metastasis rate.3 As a result, clarifying the underlying mechanism of HCC development and progression is urgently needed.
The hepatitis B virus X-interacting protein (HBXIP), encoding an 18 kDa protein, was originally identified by its interaction with the C-terminus of the hepatitis B virus X protein (HBx).4 Previous studies demonstrated that HBXIP expression was significantly increased in several cancers, such as pancreatic ductal adenocarcinomas, breast cancer, esophageal squamous cell carcinoma, lung cancer and ovarian cancer, and upregulation of HBXIP predicted poor prognosis of these patients.5–9 HBXIP mainly acts as an oncogenic transcriptional coactivator of multiple transcription factors to regulate different target genes. For instance, in breast cancer, HBXIP enhances cell proliferation and migration. HBXIP interacts with SP1 to activate the transcription of some oncogenes, including ZEB1, ACSL1, FGF4, LMO4 and PDGFB.10–14 HBXIP associates with transcription factor E2F1 to activate PKM2 and SKP2 expression.15,16 HBXIP upregulates FGF8 and VEGF to facilitate angiogenesis through inhibiting miRNA-503, which directly targeted 3 untranslated region (3ʹUTR) of FGF8 or VEGF mRNA.17 HBXIP also forms transcriptional complex with c-myc through the leucine zippers and recruits the long noncoding RNA (lncRNA) HOTAIR along with the histone demethylase LSD1 to upregulate cyclin A, eIF4E and LDHA expression.18 In HCC, HBXIP-induced activation of transcription factor E2F1 modulates SCG3 expression and facilitates proliferation.15,19 HBXIP upregulates onco-protein YAP expression through co-activating the transcription factor c-Myb, and promotes HCC growth.20 Additionally, HBXIP is able to depress the gluconeogenesis in HCC. HBXIP inhibits PCK1 expression, a key enzyme of gluconeogenesis, through downregulating transcription factor FOXO1 in HCC cells, and upregulates miR-135a targeting the 3UTR of FOXO1 mRNA. HBXIP activats PI3K/Akt pathway to induce the export of FOXO1 from nucleus to cytoplasm.21 Moreover, recently, HBXIP was found to be able to interact with HCC-associated lncRNA, lncRNA-HEIH and lncRNA-HULC.22 However, whether HBXIP have an effect on HCC metastasis and the underlying molecular mechanism is ill-documented.
Matrix metalloproteinases (MMPs) belong to a family of zinc-dependent proteolytic enzymes that are known to play a critical role in the catabolic turnover of extracellular matrix (ECM) components. MMPs are significant in the physiological processes of embryo development, wound healing and differentiation related to tissue remodeling.23,24 The elevated expression of several MMPs is involved in caner progression.25 For example, upregulation of MMP2 and MMP9 is often detected in solider tumor tissues and is associated with tumor metastasis in many cancers including HCC.26,27 MMP-9 also facilitates HBV replication through repressing IFN/JAK/STAT signaling, IFNAR1 function and IFN-α action.28 Interestingly, expression of MMPs, such as MMP2, MMP3, MMP9 and MMP13, is also observed in experimental acute and chronic liver damage.29 However, to date, the relationship between HBXIP and MMPs family remains unclear.
In this study, we aim to identify the role of HBXIP in HCC metastasis and reveal the underlying mechanism. Notably, we found that the HBXIP facilitates migration and invasion in vitro and pulmonary metastasis in vivo through upregulation of MMP15 expression in HCC. Thus, our finding provides new insights into the mechanism of HCC metastasis.
Materials and methods
Cell culture
SMMC-7721 and Bel-7402 HCC cell lines were obtained from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Hyclone) supplemented with 10% fetal bovine serum (PAN) at 37°C in 5% CO2 and 1% penicillin-streptomycin (Hyclone).
Tissue samples
Sixty-one pairs of HCC tissues and adjacent noncancerous tissues were collected from patients who initially underwent hepatectomy and were diagnosed with HCC between November 2014 and March 2016 at Zhongshan Hospital, Xiamen University. The follow-up period was up to June 2018. All subjects provided written informed consent. No patients were treated with chemotherapy and radiotherapy before hepatectomy. This study was approved by the ethics committee of the Zhongshan Hospital of Xiamen University. The research has been carried out in accordance with the World Medical Association Declaration of Helsinki.
Lentivirus-mediated gene transfer or RNA interference
The lentiviral pBobi vector was used to overexpress HBXIP. The cDNA encoding HBXIP was PCR-amplified by the Pfu Ultra II Fusion HS DNA Polymerase (Stratagene, Agilent Technologies) and subcloned into pBobi vector. Lentiviral pLKO.1 vector was used to express short hairpin RNA (shRNA) directed against HBXIP, MMP15 or c-myc. The shRNA sequences were designed by online tool (
Luciferase reporter assay
MMP15 promoter region (−1000~-1 region of MMP15 gene) was cloned into pGL3-basic reporter vector (pGL3-MMP15). Cells were cotransfected with pGL3-basic or pGL3-MMP15 and pRL-TK. After 24 hrs, luciferase activity was determined using Dual-Luciferase® Reporter Assay System (Promega), according to the manufacturer’s instructions. In brief, cells were lysed using Passive Lysis Buffer. After 15 mins, carefully transfer up to 20 µL of cell lysate into the luminometer tube containing LAR II; mix by pipetting 2 or 3 times. Place the tube in the luminometer and initiate reading. Then, add 100 µL of Stop & Glo® Reagent and vortex briefly to mix. Replace the sample in the luminometer, and initiate reading. Relative luciferase activity was detected using Varioskan Lux detection System (Thermo Scientific) and was normalized to Renilla luciferase activity at 48 hrs after transfection.
Chromatin immunoprecipitation (ChIP) assays
ChIP assay was performed using an EZ-Magna ChIP kit according to its manufacturer’s instruction. ChIP-enriched promoter of MMP15 genes was quantified by qRT-PCR using the following primers (cover −349~-282): 5ʹ- TGAACCGTGTCCCTCCTAAA −3ʹ, 5ʹ- GGTGTCTCTGGCCATTGAATAA −3ʹ. In brief, cells were cross-linked in 1% formaldehyde at room temperature for 10 mins. Crosslinking was terminated by the addition of 125 mM glycine. Cell pellets were lysed using cell lysis buffer and then centrifuged at 5000 rpm for 5 mins at 4°C to pellet the nuclei. Pellets were lysed using nuclear lysis buffer and then diluted with IP dilution buffer. Nuclear lysates were sonicated and the debris was removed by centrifugation. HBXIP antibodies (Abcam) antibodies or IgG antibodies (Millipore) were mixed with clear nuclear lysates for immunoprecipitation. Coprecipitated DNA was purified and the level of target genes was quantified using real-time PCR.
Western blot
Cells and tissues were lysed in RIPA lysis buffer (Beyotime, Beijing, China) containing the protease inhibitor phenylmethanesulfonyl fluoride (PMSF). Protein lysates were subjected to SDS-PAGE and transferred to PVDF membranes (Millipore). Membranes were incubated with anti-HBXIP (Santa Cruz, 1:300), anti-MMP15 (Thermo,1:1,000) or anti-β-actin (Cell Signaling Technology, 1:2,000) antibodies at 4°C overnight, and followed by incubation with secondary antibodies conjugated with horseradish peroxidase (HRP, Jackson). Immunoreactive proteins were visualized using the ECL detection system (Millipore).
RNA isolation and quantitative real-time PCR (qrt-PCR) analyses
Total RNA was isolated by TRIzol reagent (Invitrogen) according to standard protocol. cDNA was synthesized using One-Step gDNA Removal and cDNA Synthesis Kit (Transgen, Beijing, China). Real-time PCR was performed in the Lightcycle™ Real-Time PCR System (Roche) using FastStart Universal SYBR Green Master (Rox) (Roche). Relative expression levels were calculated as ratios normalized against those of ACTB. Comparative quantification was determined using the 2–ΔΔCt.
Migration and invasion assays
24-well Transwell chambers with 8 µm pore size polycarbonate membrane were used (Corning Inc., Corning, NY, USA) for cell migration and invasion assays. For migration assay, 1.0×105 cells were planted on the top side of the membrane. For invasion assay, 1.0×105 cells were planted on the top side of the membrane precoated with Matrigel (BD Bioscience). After 24 hrs, the cells that had passed through the membrane to the lower surface were fixed with methanol, stained with crystal violet, and then counted.
Animal studies
A xenograft mouse model was developed using 6-week-old male BALB/c athymic nude mice. Stable HCC cells with HBXIP alteration were trypsinized and harvested in serum-free DMEM, and 0.1 mL serum-free DMEM containing 5×106 cells was injected subcutaneously into the right flank of the nude mice. After 40 days, mice were sacrificed by breaking the neck to death, and the tumors and lung tissues were removed for further analysis. All animal experiments were approved by the Animal Care and Use Committee of the Xiamen University according to the Guidelines Followed for the Welfare of the Animals of the Xiamen University. To detect pulmonary metastasis, all the lung tissues were fixed in 10% neutral formalin, embedded in paraffin, and cut into 3-μm-thick sections. Hematoxylin and eosin (HE) staining was performed in all lung tissues using a standard protocol. Then, the total number of pulmonary metastasis in every mouse was counted under a microscope (Leica, Germany). One metastasis nodule contains more than fifty HCC cells. We counted all the metastasis nodules in all lung tissues of each mice and compared the difference between different groups.
Statistical analyses
All statistical analyses in our experiment were performed using the SPSS version 20.0 software (IBM, Armonk, NY, USA). All experiments were performed in triplicate. Data are shown as mean ± standard error of mean (SEM). The differences between two groups were analyzed by the Student’s t-test or Chi-square test. Paired student’s t-test was used to analyse the differential expression of HBXIP between HCC and corresponding nontumor tissue samples. The Chi-square test was performed for analyzing the correlation between HBXIP expression and clinicopathological features of HCC patients. ANOVA and LSD test was used to make comparisons of multiple groups. The Kaplan–Meier method and log-rank test was performed for analyzing the correlation between HBXIP expression and overall survival rate of patients with HCC. The Pearson correlation analysis was performed for analyzing the correlation between HBXIP and MMP15 mRNA expression in 35 HCC tissues. P<0.05 was considered statistically significant difference.
Results
HBXIP is upregulated in HCC and predicts poor prognosis of patients with HCC
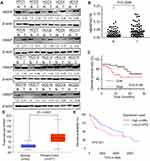
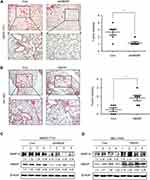
Firstly, we analyzed the HBXIP protein level of HCC and correspondence nontumor tissues from 20 patients with HCC. 65% (13/20) HCC tissues had higher level of HBXIP protein expression than nontumor tissues (Figure 1A). We also detected the HBXIP mRNA expression in 61 pairs of HCC and corresponding nontumor tissue samples by qRT-PCR. As shown in Figure 1B, the HBXIP expression was significantly increased in 68.9% (42/61) HCC tissues compared with that in matched nontumor tissues. To reveal the clinical significance of HBXIP expression, we analyzed the correlation between HBXIP mRNA levels and clinicopathological characteristics of HCC patients. The median value of HBXIP mRNA expression in HCC tissues was set as the threshold value, and the tissue samples were divided into high-level group (n=31) and low-level group (n=30). It was found that upregulation of HBXIP expression was significantly associated with vascular invasion and tumor metastasis (P=0.029) (Table 1). However, HBXIP expression was not associated with gender, age, HbsAg, AFP, tumor size, liver cirrhosis and differentiation level. Moreover, the Kaplan–Meier survival analysis demonstrated that high level of HBXIP mRNA expression was associated with poor overall survival in HCC patients (Figure 1C). Moreover, we analyzed the HCC database of The Cancer Genome Atlas (TCGA) to evaluate the differential expression of HBXIP mRNA between normal and HCC tissues by using online tool, UALCAN analysis (
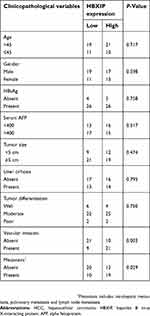 | Table 1 Correlation between HBXIP expression and clinicopathological features of HCC patients |
HBXIP promotes the migration and invasion of HCC cells
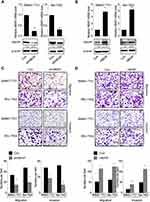
The positive correlation between HBXIP expression and metastasis implied that HBXIP may promote the metastasis of HCC cells. To evaluate the effects of HBXIP on migration and invasion in vitro, we constructed cell lines with HBXIP stable overexpression and knockdown. Both the upregulation and knockdown of HBXIP expression were confirmed by western blotting and qRT-PCR analysis (Figure 2A and B). Silencing endogenous HBXIP expression markedly suppressed migration and invasion in SMMC-7721 and Bel-7402 cells (Figure 2C). Conversely, overexpression of HBXIP significantly enhanced the migration and invasion capacities of SMMC-7721 and Bel-7402 cells (Figure 2D). These data demonstrated that HBXIP enhances the migratory and invasive ability of HCC cells in vitro.
HBXIP increases MMP15 expression partly through transcription factor c-myc
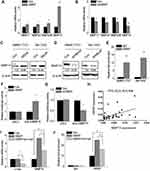
Next, we explored the underlying mechanisms by which HBXIP regulated HCC progression. A previous study found that 388 gene promoters could be occupied by both HBXIP and c-myc.18 We found that HBXIP may regulate some members of MMPs family, including MMP11, MMP15, MMP23 and MMP23B. To confirm this result, we detected the expression change of these members of MMPs family in SMMC-7721 cells with HBXIP overexpression by qRT-PCR. Notably, MMP11, MMP23 and MMP23B were slightly changed, while MMP15 was markedly increased by HBXIP upregulation (Figure 3A). In contrast, knockdown of HBXIP significantly suppressed MMP15 mRNA expression, and have only a slight effect on MMP11, MMP23 and MMP23B expression (Figure 3B). The western blot assays also demonstrated that HBXIP overexpression increased MMP15 protein level (Figure 3C), whereas HBXIP silence exerted the opposite effect (Figure 3D). In addition, a ChIP assay confirmed the binding of HBXIP in the MMP15 promoter in HCC cells (Figure 3E). A luciferase reporter assay showed that HBXIP overexpression significantly induced transactivation of the MMP15 promoter (Figure 3F), whereas HBXIP knockdown suppressed the activity of MMP15 promoter luciferase reporter vector (Figure 3G). In addition, we analyzed the pathological correlation between HBXIP and MMP15 expression. We detected the MMP15 and HBXIP expression in HCC tissues by qRT-PCR, and the correlation analyses showed a positive correlation between HBXIP and MMP15 expression in HCC tissues (Figure 3H). These results suggest that HBXIP transactivates MMP15 expression.
Previous study demonstrated that among 388 c-myc target genes, such as Cyclin A, eIF4E and LDHA, their promoters could be occupied by the oncoprotein HBXIP. Knockdown of HBXIP abolished c-myc–mediated upregulation of these target genes. Mechanistically, HBXIP directly interacted with c-myc through the leucine zippers.18 We suspected that MMP15 was regulated by HBXIP in such a manner. The results showed that the silence of c-myc partly abolished the HBXIP-induced upregulation of MMP15 (Figure 3I). Moreover, a ChIP assay demonstrated that c-myc downregulation partially decreased the binding level of HBXIP to the MMP15 promoter regions, suggesting that other molecule mechanisms may be involved in HBXIP-induced upregulation of MMP15 (Figure 3J). Taken together, our data suggest that HBXIP activates MMP15 transcription partially through c-myc.
HBXIP promotes HCC cell migration and invasion partly through MMP15
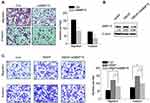
As shown in Figure 4A, knockdown of MMP15 significantly decreased migration and invasion in SMMC-7721 cells. To examine whether the effect of HBXIP on HCC cell migration and invasion depends on MMP15, we silenced MMP15 expression in SMMC-7721 cells with HBXIP overexpression (Figure 4B). Downregulation of MMP15 partially decreased the cell migration and invasion mediated by HBXIP overexpression (Figure 4C). These findings suggest that HBXIP promotes cell migration and invasion partly through activation of MMP15.
HBXIP facilitates the metastasis of HCC cells in vivo
To further explore the role of HBXIP in tumor metastasis in vivo, HCC cells were subcutaneously transplanted into nude mice. Histological analysis confirmed that the number of lung metastatic nodules in the SMMC-7721-shHBXIP group was significantly reduced, compared to that in the SMMC-7721-control group (Figure 5A); however, the number of lung metastatic nodules in the Bel-7402-HBXIP group was increased, compared to that in the Bel-7402-control group (Figure 5B). Moreover, xenografts in nude mice with SMMC-7721-shHBXIP expressed lower MMP15 (Figure 5C). Conversely, the MMP15 expression was significantly increased in Bel-7402-HBXIP xenografts compared to control group (Figure 5D). Thus, we concluded that HBXIP promotes the metastasis of HCC cells via MMP15 in vivo.
Discussion
Previous studies demonstrated that HBXIP expression was upregulated in some cancers, such as breast cancer, lung cancer and ovarian cancer. HBXIP can be a predictor for clinical outcomes of patients with cancer.5–7,30 Even though HBXIP was firstly identified in HCC, the expression pattern and clinical significance in HCC remains unclear. In the present study, we found a significant increase of HBXIP mRNA and protein level in HCC tissues. HBXIP overexpression was correlated with vascular invasion and tumor metastasis. In addition, HCC patients with high-level HBXIP expression had worse prognoses than did patients with low-level HBXIP expression. Our clinical data suggested that overexpression of HBXIP contributes to the progression of HCC and may be a potential biomarker for prognosis.
Recurrence and metastasis is the most common lethal cause after surgical resection and adjuvant therapy in HCC.31 Metastatic disease is largely incurable because of its systemic nature and the resistance of disseminated tumor cells to the therapeutic agents.32 Thus, it is urgently needed to investigate the molecular mechanisms underlying HCC metastasis. Active proteolysis effected principally by MMPs, drives the loss of the basement membrane (BM) barrier, which allows direct invasion by cancer cells of the stromal compartment. In normal cells, the activity of MMPs is carefully regulated by transcriptional or posttranslational mechanisms. However, cancer cells have numerous ways by which to derail the normally tight control of MMPs activity, almost leading to enhanced MMPs function.33 Our present study showed that HBXIP enhanced the cellular migration and invasion of HCC cells in vitro, and promoted pulmonary metastasis in vivo. Mechanistically, ChIP and luciferase assays demonstrated that HBXIP bind to MMP15 promoter region, and increase the activity of MMP15 promoter. Moreover, the positive correlation between HBXIP and MMP15 expression further supported that MMP15 was a downstream target gene of HBXIP. Further experiments, such as electrophoretic mobility shift assay (EMSA) and DNA pull-down assay, should be performed to validate whether HBXIP directly bind to the promoter region of MMP15.
c-myc, a basic-helix-loop-helix-leucine zipper protein, plays critical roles in multiple cellular processes that sustain growth of many types of cancers. The target genes of c-myc participate in many cellular processes, including cell cycle, apoptosis, protein synthesis, migration, invasion and noncoding RNA expression.34 Recently, HBXIP was found to function as a transcriptional co-activator of c-myc to activate cyclin A, eIF4E and LDHA.18 Based on this finding, we also found that HBXIP upregulated MMP15 expression through association with c-myc. Knockdown of c-myc attenuated the occupancy of HBXIP in MMP15 promoter, and subsequently abolished the HBXIP-induced MMP15 upregulation. However, c-myc silence could not completely abolish the MMP15 increased by HBXIP. Other molecular mechanisms may be involved in HBXIP-mediated HCC metastasis, which needs further investigation.
Conclusion
Collectively, our study reveals that HBXIP upregulation in HCC is a strong indicator of more-aggressive tumors and poor prognosis. HBXIP facilitates HCC metastasis through activation of MMP15 transcription. Thus, HBXIP may be a candidate biomarker for HCC prognosis and a therapeutic target for HCC treatment.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No.81672418, 81702351, 81871961, 81702348, 81572335 and 81602500), the Natural Science Foundation of Fujian (No.2017-2-101, 2018J01389, 2018J01398, 2016-2-80), Research Project of health and family planning (no. 2018-2-64 and 2018-ZQN-84) and the Science and Technology Project of Xiamen (No. 3502Z20164023, 3502Z20164022).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–399. doi:10.3322/caac.21161
3. Goh GB, Chang PE, Tan CK. Changing epidemiology of hepatocellular carcinoma in Asia. Best Pract Res Clin Gastroenterol. 2015;29(6):919–928. doi:10.1016/j.bpg.2015.09.007
4. Melegari M, Scaglioni PP, Wands JR. Cloning and characterization of a novel hepatitis B virus x binding protein that inhibits viral replication. J Virol. 1998;72(3):1737–1743.
5. Wang Y, Li N, Che S, et al. HBXIP suppression reduces cell proliferation and migration and its overexpression predicts poor prognosis in non-small-cell lung cancer. Tumour Biol. 2017;39(7):1010428317709675. doi:10.1177/1010428317709675
6. Wang Y, Sun J, Li N, et al. HBXIP overexpression is correlated with the clinical features and survival outcome of ovarian cancer. J Ovarian Res. 2017;10(1):26. doi:10.1186/s13048-017-0322-7
7. Cheng D, Liang B, Li Y. HBXIP expression predicts patient prognosis in breast cancer. Med Oncol. 2014;31(10):210. doi:10.1007/s12032-014-0374-0
8. Zhou X, Wang X, Duan J, et al. HBXIP protein overexpression predicts the poor prognosis of pancreatic ductal adenocarcinomas. Pathol Res Pract. 2019;215(2):343–346. doi:10.1016/j.prp.2018.12.016
9. Xia H, Ma L, Li J, Bai H, Wang D. Elevated HBXIP expression is associated with aggressive phenotype and poor prognosis in esophageal squamous cell carcinoma. Am J Cancer Res. 2017;7(11):2190–2198.
10. Jiang Y, Wang D, Ren H, Shi Y, Gao Y. Oncogenic HBXIP enhances ZEB1 through Sp1 to accelerate breast cancer growth. Thorc Cancer. 2018;9(12):1664–1670. doi:10.1111/1759-7714.12878
11. Yue L, Li L, Liu F, et al. The oncoprotein HBXIP activates transcriptional coregulatory protein LMO4 via Sp1 to promote proliferation of breast cancer cells. Carcinogenesis. 2013;34(4):927–935. doi:10.1093/carcin/bgs399
12. Zhang Y, Zhao Y, Li L, et al. The oncoprotein HBXIP upregulates PDGFB via activating transcription factor Sp1 to promote the proliferation of breast cancer cells. Biochem Biophys Res Commun. 2013;434(2):305–310. doi:10.1016/j.bbrc.2013.02.123
13. Shi H, Li Y, Feng G, et al. The oncoprotein HBXIP up-regulates FGF4 through activating transcriptional factor Sp1 to promote the migration of breast cancer cells. Biochem Biophys Res Commun. 2016;471(1):89–94. doi:10.1016/j.bbrc.2016.01.174
14. Wang Y, Cai X, Zhang S, et al. HBXIP up-regulates ACSL1 through activating transcriptional factor Sp1 in breast cancer. Biochem Biophys Res Commun. 2017;484(3):565–571. doi:10.1016/j.bbrc.2017.01.126
15. Xu F, You X, Liu F, et al. The oncoprotein HBXIP up-regulates Skp2 via activating transcription factor E2F1 to promote proliferation of breast cancer cells. Cancer Lett. 2013;333(1):124–132. doi:10.1016/j.canlet.2013.01.029
16. Liu BW, Wang TJ, Li LL, et al. Oncoprotein HBXIP induces PKM2 via transcription factor E2F1 to promote cell proliferation in ER-positive breast cancer. Acta Pharmacol Sin. 2019 Apr;40(4):530–538.
17. Liu F, You X, Wang Y, et al. The oncoprotein HBXIP enhances angiogenesis and growth of breast cancer through modulating FGF8 and VEGF. Carcinogenesis. 2014;35(5):1144–1153. doi:10.1093/carcin/bgu021
18. Li Y, Wang Z, Shi H, et al. HBXIP and LSD1 scaffolded by lncRNA hotair mediate transcriptional activation by c-Myc. Cancer Res. 2016;76(2):293–304. doi:10.1158/0008-5472.CAN-14-3607
19. Wang Y, Cui M, Cai X, et al. The oncoprotein HBXIP up-regulates SCG3 through modulating E2F1 and miR-509-3p in hepatoma cells. Cancer Lett. 2014;352(2):169–178. doi:10.1016/j.canlet.2014.05.007
20. Wang Y, Fang R, Cui M, et al. The oncoprotein HBXIP up-regulates YAP through activation of transcription factor c-Myb to promote growth of liver cancer. Cancer Lett. 2017;385:234–242. doi:10.1016/j.canlet.2016.10.018
21. Shi H, Fang R, Li Y, et al. The oncoprotein HBXIP suppresses gluconeogenesis through modulating PCK1 to enhance the growth of hepatoma cells. Cancer Lett. 2016;382(2):147–156. doi:10.1016/j.canlet.2016.08.025
22. Ruan L, Huang L, Zhao L, et al. The interaction of lncRNA-HEIH and lncRNA-HULC with HBXIP in Hepatitis B Patients. Gastroenterol Res Pract. 2018;2018:9187316. doi:10.1155/2018/9187316
23. Haas CS, Gleason B, Lin S, Tramonti G, Kanwar YS. Matrix metalloproteinases in renal development. Connect Tissue Res. 2004;45(2):73–85. doi:10.1080/03008200490442644
24. Tomlinson ML, Garcia-Morales C, Abu-Elmagd M, Wheeler GN. Three matrix metalloproteinases are required in vivo for macrophage migration during embryonic development. Mech Dev. 2008;125(11–12):1059–1070. doi:10.1016/j.mod.2008.07.005
25. Maciejczyk M, Pietrzykowska A, Zalewska A, Knas M, Daniszewska I. The significance of matrix metalloproteinases in oral diseases. Adv Clin Exp Med. 2016;25(2):383–390. doi:10.17219/acem/30428
26. Zhang M, Dai C, Zhu H, et al. Cyclophilin A promotes human hepatocellular carcinoma cell metastasis via regulation of MMP3 and MMP9. Mol Cell Biochem. 2011;357(1–2):387–395. doi:10.1007/s11010-011-0909-z
27. Huang D, Du X, Yuan R, et al. Rock2 promotes the invasion and metastasis of hepatocellular carcinoma by modifying MMP2 ubiquitination and degradation. Biochem Biophys Res Commun. 2014;453(1):49–56. doi:10.1016/j.bbrc.2014.09.061
28. Chen J, Xu W, Chen Y, et al. Matrix metalloproteinase 9 facilitates hepatitis B virus replication through binding with type i interferon (IFN) receptor 1 to repress IFN/JAK/STAT signaling. J Virol. 2017;91(8). doi:10.1128/JVI.00955-17
29. Hemmann S, Graf J, Roderfeld M, Roeb E. Expression of MMPs and TIMPs in liver fibrosis – a systematic review with special emphasis on anti-fibrotic strategies. J Hepatol. 2007;46(5):955–975. doi:10.1016/j.jhep.2007.02.003
30. Zhao Y, Li H, Zhang Y, et al. Oncoprotein HBXIP modulates abnormal lipid metabolism and growth of breast cancer cells by activating the LXRs/SREBP-1c/FAS signaling cascade. Cancer Res. 2016;76(16):4696–4707. doi:10.1158/0008-5472.CAN-15-1734
31. El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365(12):1118–1127. doi:10.1056/NEJMra1001683
32. Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi:10.1016/j.cell.2006.11.001
33. Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi:10.1016/j.cell.2010.03.015
34. Sipos F, Firneisz G, Muzes G. Therapeutic aspects of c-MYC signaling in inflammatory and cancerous colonic diseases. World J Gastroenterol. 2016;22(35):7938–7950. doi:10.3748/wjg.v22.i35.7938
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.