Back to Journals » Cancer Management and Research » Volume 12
The Novel Zinc Finger Protein 587B Gene, ZNF587B, Regulates Cell Proliferation and Metastasis in Ovarian Cancer Cells in vivo and in vitro
Authors Liu Y, Ouyang Q, Sun Z, Tan J, Huang W, Liu J, Liu Z, Zhou H, Zeng F, Liu Y
Received 3 March 2020
Accepted for publication 2 June 2020
Published 26 June 2020 Volume 2020:12 Pages 5119—5130
DOI https://doi.org/10.2147/CMAR.S252347
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Ahmet Emre Eşkazan
Yujie Liu,1,2 Qianying Ouyang,1,2 Zeen Sun,1,2 Jieqiong Tan,3 Weihua Huang,1,2 Jie Liu,1,2 Zhaoqian Liu,1,2 Honghao Zhou,1,2 Feiyue Zeng,4,* Yingzi Liu1,2,*
1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410008, People’s Republic of China; 2Institute of Clinical Pharmacology, Central South University, Hunan Key Laboratory of Pharmacogenetics, Changsha 410078, People’s Republic of China; 3National Laboratory of Medical Genetics, Central South University, Changsha 410078, People’s Republic of China; 4Department of Radiology, Xiangya Hospital, Central South University, Changsha 410008, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yingzi Liu
Institute of Clinical Pharmacology, Central South University; Hunan Key Laboratory of Pharmacogenetics, Changsha 410078, People’s Republic of China
Tel +86 731 84805380
Fax +86 731 82354476
Email [email protected]
Feiyue Zeng
Department of Radiology, Xiangya Hospital, Central South University, Changsha 410008, People’s Republic of China
Tel +86 731 84805380
Fax +86 731 82354476
Email [email protected]
Background: The zinc finger protein 587B (ZNF587B) is a novel cisplatin-sensitive gene that was identified in our previous research by using a genome-scale CRISPR-Cas9 knockout library in ovarian cancer (OC) cell lines. ZNF587B belongs to the C2H2-type zinc finger protein (ZFP) family. Many ZFP protein could inhibit tumor development and malignancy. However, the function of ZNF587B remains unknown.
Methods: Quantitative PCR (qPCR) was utilized to compare ZNF587B mRNA expression levels in OC and normal ovarian cell lines. The small interfering RNA (siRNA) and full-length ZNF587B eukaryotic expression plasmid were constructed and transfected into OC cells later. Colony formation, 5-ethynyl-2′-deoxyuridine (EdU) assay, transwell assay, and xenograft experiment were conducted to evaluate the effect of ZNF587B on OC cells.
Results: ZNF587B was downregulated by approximately 43% and 17% in the OC cell lines SKOV3 and A2780, respectively, compared with that in the normal ovarian cell line IOSE80. Overexpression of ZNF587B reduced cell proliferation, colony formation, migration, and invasion, which could be reversed by knockdown of ZNF587B via siRNA. Xenograft experiments also confirmed that ZNF587B could suppress tumor growth. Survival data of OC patients in the SurvExpress database showed that with respect to overall survival, low-risk patients grouped by the prognostic index had a higher expression of ZNF587B and a better prognosis than high-risk group (HR = 1.77, 95% CI: 0.55– 0.70, p = 0.023). Moreover, overexpression of ZNF587B promoted OC cells apoptosis when pretreated with cisplatin.
Conclusion: ZNF587B is a novel potential tumor suppressor of OC and may be a therapeutic target for OC.
Keywords: ZNF587B, C2H2-type zinc finger protein, proliferation, metastasis, ovarian cancer
Introduction
Ovarian cancer (OC) is the leading cause of mortality among common gynecological malignancies.1 More than 70% of patients are diagnosed at an advanced stage III or IV (International Federation of Gynecology and Obstetrics, FIGO) with distant metastases.2–4 The standard therapy for OC is platinum-based chemotherapy following cytoreductive surgery.5 Generally, most patients have a high initial response rate to cisplatin. Unfortunately, more than 60% of patients relapse within 18 months, and almost all patients relapse within 3 years due to drug resistance.6,7 The 10-year progression-free survival rate of recurrent patients is less than 15%.2 Over the past 50 years, many drug resistance associated genes such as CTR1, GSTP1, ERCC1, XIAP and NT5E,8–12 involving in drug transport, DNA repair, and drug transformation had been intensively studied.13–16 However, no powerful genes have been identified so far. Recently, the clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 approach has become a biotechnology breakthrough due to its high knockout efficiency, specificity, and reproducibility,17–19 and has been widely applied to cancer research and medicine. We employed the whole human genome-scale CRISPR-Cas9 knockout library to screen genes for cisplatin resistance in OC and found that the ZNF587B could facilitate cisplatin-induced cytotoxicity,20 and would serve as a novel potential biomarker for predicting cisplatin resistance. To date, few pertinent studies about ZNF587B have been reported.
ZNF587B, as a member of the Krüppel-type zinc-finger proteins (KRAB-ZFPs) family, may have a KRAB domain and six tandems of C2H2-type zinc finger motifs according to SMART database (http://smart.embl.de). The KRAB domain is one of the most widely distributed and potent transcriptional repressor domains, whereas the C2H2 zinc finger motif binds DNA.21–28 KRAB-ZFPs probably constitute the single-largest class of transcription factors within the human genome. Although their function is largely unknown, emerging evidences demonstrate they play important roles in cell proliferation, apoptosis, cancer, and chemoresistance.21,29,30 For instance, Wei et al found that the ZFP Egr-1 was downregulated in breast cancer tissues versus benign tissues, and suppressed cell proliferation.31 In non-small-cell lung carcinoma, Egr-1 inhibited cell mobility and induced apoptosis.32 The expression of another ZFP, ZFP403, was lower in OC tissues than normal ovarian tissues, and this gene inhibited cell proliferation and ovarian tumor growth.33 However, little is known about ZNF587B. In our previous study, we reported that ZNF587B was related to cisplatin resistance in OC. Herein, we sought to explore its role in OC.
Materials and Methods
Cell Culture
The normal ovarian cell line IOSE80 was purchased from American Type Culture Collection (ATCC, USA) and cultured in RPMI-1640 medium (HyClone, USA) with 10% fetal bovine serum (FBS, [Biological Industries, Israel]). The human ovarian carcinoma cell lines A2780 and SKOV3 were obtained from ATCC and cultured in DMEM medium and McCoy’s 5A medium (Biological Industries, Israel) containing 10% FBS, respectively. Cells were cultivated at 37°C in a humidified atmosphere containing 5% CO2.
Plasmids, Small Interfering RNA (siRNA) Transfection and Generation of Stable Cell Lines
The siRNA for ZNF587B with the sequence 5′-GTTCAAACGTGAACCTTAA-3′ was synthesized by RiboBio (China). This sequence was obtained from our previous studies, and was shown to have the lowest knockdown efficiency in OC lines.20 The expression plasmid pcDNA3.1-ZNF587B with the Flag epitope was constructed by Genechem (China). Transfections were performed in A2780 cells with Lipofectamine 2000 reagent (Thermo Fisher, USA) according to the manufacturer’s protocol. After transfection, cells were screened with 500 μg/mL G418 (Beyotime, China) for 15 days. Monoclonal cells were transferred into 35 mm culture dishes, and A2780 cells stably expressing ZNF587B were expanded in medium containing G418. Quantitative polymerase chain reaction (qPCR) was performed to verify stable transfections after 30 generations of cell culture.
Cell Proliferation and Clone Formation
Cells were resuspended in 6-well plates in complete medium with 10% FBS and transfected with either siRNA or plasmid for 48 h. Cell proliferation in vitro was assayed by EdU (5-Ethynyl-2′-deoxyuridine), a thymidine analog that is incorporated into DNA during the proliferative phase. Fluorescence microscopy was used to detect EdU via Apollo®-based fluorescent dyes. For the plate colony formation assay, an equal number of cells containing either siRNA-ZNF587B or pcDNA3.1-ZNF587B along with their respective control groups were seeded in 6-well plates (500 cells/well) and cultured for 14 days. The culture medium was changed every three days. On the final day, cells were fixed with 4% paraformaldehyde and stained with freshly prepared crystal violet for 30 min. Colony formation was observed by microscopy. Experiments were repeated a minimum of three times and reported as the mean ± standard deviation (SD).
Migration and Invasion Assays
Transwell chambers (8 μm, Corning, NY) without Matrigel was used to measure cell migration. For invasion potential assays, 80 μL of 1:8 Matrigel and FBS-free medium was added to the bottom of the chamber. After 6 h in cell incubator, 200 μL of cell suspension (approximately 1 × 104 cells) with FBS-free medium was added to the upper chamber, and 600 μL of complete medium with 10% FBS was added to the lower chamber. After 48 h, migrated or invasive cells were fixed with 4% paraformaldehyde for 30 min and stained with 0.1% crystal violet for 1 h. The number of stained cells was calculated in 3 random fields using an optical microscope. Experiments were repeated a minimum of three times and reported as the mean ± SD.
RNA Isolation and Quantitative PCR
Total RNA was extracted from cultured cells using Trizol reagent (Takara, Japan) according to the manufacturer’s protocol. Approximately 1 μg of RNA was reverse transcribed using the Prime Script RT reagent kit with gDNA Eraser (Takara, Japan), and the qPCR was performed on a Roche thermocycler (Roche) using 2×SYBR Green qPCR Master Mix (Bimake.cn China). The mRNA expression levels of ZNF587B were determined by normalizing to ACTB mRNA expression. The primers used for qPCR were as follows (BioSune, China): ZNF587B Fw: GCGCCATCAAAAAGTTCACG; Rev: GCTGGGCTTCCGACTAAAAG. Data were analyzed using the 2−Δct method. All experiments were performed a minimum of three times.
CCK-8 Assay
Cells were seeded in 96-well plates at a density of 1 × 105 L after infection for 48 h and incubated with a series of concentrations of cisplatin (Sigma-Aldrich, USA) for another 48 h. Chemosensitivity was determined to evaluate the cell viability by CCK-8 approach (Takara, Japan) according to protocol. Cell survival and IC50 were calculated from the dose response curves using GraphPad Prism 7 (GraphPad software, CA). All experiments were performed in triplicate.
Flow Cytometric Analysis of Apoptosis
The apoptosis-inducing effect of cisplatin in OC cell lines was determined using a PE Annexin V/7-AAD apoptosis detection kit (BD Pharmingen, USA). A2780 cells were seeded in 6-well plates at a number of 2×105 cells/well. After transfection with either siRNA or plasmid for 48 h, cells were cultured with the IC50 concentration of cisplatin for another 48 h. Adhering and floating cells were subsequently collected by trypsinization and centrifugation at 3000 rpm for 5 min. After washing twice with cold PBS, cells were diluted in annexin binding buffer at a concentration of 1×106 L. Five microliters of PE Annexin V/7-AAD was added and the cells were incubated for 15 minutes at room temperature in the dark according to the manufacturer’s protocol. Stained cells were analyzed in different experimental groups using a flow cytometer (FACSCalibur, BD Biosciences).
SurvExpress Database Analysis
We analyzed data from the OV-AU-ICGC OC serous cystadenocarcinoma dataset generated by the International Cancer Genome Consortium (ICGC) on the SurvExpress database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). This database is a comprehensive cancer-wide gene expression platform that includes clinical outcomes and survival analyses of cancer datasets. High-/low-risk groups were determined using an algorithm of the prognostic risk score. Data were input for Cox proportional hazards regression using a list of biomarker genes of interest for risk estimation.
In vivo Cell Line-Derived Xenograft Study
All animal care and experimental procedures were conducted according to the National Institutes of Health’s Guide for the Use and Care of Laboratory Animals, and approved by the Institutional Review Board of Central South University (Changsha, China). Female BALB/c nude mice (4 weeks of age, 16–20 g) were obtained from Hunan SJA Laboratory Animal Co., Ltd, and were fed sterilized food and water under specific pathogen-free conditions at the Department of Laboratory Animals, Central South University. The mice were then randomly divided into two groups of three each. Mice were inoculated subcutaneously into both sides of the flank with 1 x 107 cells of ZNF587B stable transfection A2780 cell line or the corresponding vector control in serum-free medium. The tumors were examined every 2 days, and the maximum (L) and minimum (W) lengths of the tumors were measured with calipers. Tumor volumes were calculated as 0.5 LW.2 Mice were sacrificed by intraperitoneal injection with 200 μL of 10% chloral hydrate when the tumor volume was 200–300 mm3. Tumors were excised after the skin was disinfected, placed in approximately 1.5 mm3 normal saline, inoculated into the animal’s bilateral armpit with a trocar, and the skin wound was disinfected with iodophor. The tumors were examined every 2 days. On day 24, the animals were euthanized, and the tumors were excised and weighed.
Statistical Analysis
For parametric data, comparisons between two groups were performed using the 2-tailed Student’s t-test. All data were reported as the mean ± SD. All statistical analyses were performed using SPSS 22.0 or GraphPad Prism 7. In all cases, differences were considered statistically significant at P < 0.05.
Results
ZNF587B Is Downregulated in Human OC Cells
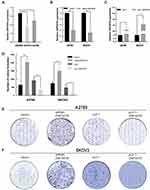
To determine the differences in ZNF587B expression between OC cell lines and ovarian normal cell line, qPCR analyses were performed to compare mRNA expression levels in two OC cell lines (SKOV3 and A2780) and a normal ovarian epithelial cell line (IOSE80). As shown in Figure 1A, the expression of ZNF587B decreased approximately 43% and 17% in SKOV3 and A2780 cells, respectively, compared with that in IOSE80 cells (P < 0.01, P < 0.05, respectively). The expression of ZNF587B was lowest in SKOV3 cells. These results showed that the expression of ZNF587B in OC cell lines was significantly decreased compared with that in normal ovarian cell lines.
ZNF587B and Cell Proliferation
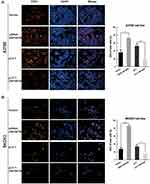
To investigate the effects of ZNF587B on proliferation, clone formation and EdU assays were carried out in OC cell lines. A2780 and SKOV3 cell lines were transfected with either ZNF587B siRNA or full-length expression plasmid pcDNA3.1-ZNF587B to alter the expression of ZNF587B. Expression levels were confirmed by qPCR, as shown in Figure 1B–C. Larger colony sizes, greater colony numbers (P < 0.05), and a larger number of EdU−positive cells (P < 0.01) were observed in the siRNA-treated groups than in the vector control groups (Figure 1D–F; Figure 2A and B). In contrast, overexpression of ZNF587B impaired cell clonality and proliferation (Figure 1D–F and 2a-b; P < 0.05, P < 0.01, respectively). In summary, our results indicated that ZNF587B could inhibit cell clonality and proliferation.
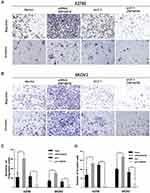
ZNF587B Inhibits Migration and Invasion of OC Cell Lines
The cell–cell adhesion of tumor cells is often reduced, which refers to with tumor metastasis and invasion. To evaluate whether ZNF587B was implicated in metastasis and invasion ability of OC cells, a Boyden chamber assay was performed on A2780 and SKOV3 cells transfected with either siRNA-ZNF587B or full-length ZNF587B plasmid. Representative migration and invasion images are shown in Figure 3. Compared with the control group, the migration cells through the polycarbonate membrane were obviously increased in siZNF587B-treated group in A2780 (2.83 times, P < 0.01) and SKOV3 (2.21 times, P < 0.001). Conversely, the migrating cell number significantly decreased by 66% and 45% in the ZNF587B overexpression groups of A2780 and SKOV3 compared with that in the control group (P < 0.05, P < 0.01, respectively, Figure 3A). Similarly, OC cellular invasive ability was also altered by variation in the expression of ZNF587B, as shown in Figure 3B. In Figure 3C and D, the histogram indicates that ZNF587B impairs cell adhesion and suppresses migration and invasion.
Ectopic Overexpression of ZNF587B Suppresses Tumor Growth in vivo
Based on the significant inhibitory effect of ZNF587B on proliferation, migration, and invasion of OC cells, we further investigated whether ZNF587B affected tumor growth in vivo. Nude mouse tumorigenicity assay was carried out to validate the effect of ZNF587B in vivo. Due to the relatively lower expression of ZNF587B in A2780 cells than in SKOV3 cells, A2780 was used for subcutaneous xenograft tumor model. An equal number of stably transfected pc3.1-ZNF587B or pc3.1 A2780 cells were injected subcutaneously into nude mice (Figure 4A). The tumors were removed on the 27th day after inoculation. No significant difference in body weight was observed between two group mice (Figure 4B), which indicated that ZNF587B had no toxicity in mice. Tumor volume and mice weight were recorded every 2 days, and tumor weight was measured on the 27th day (Figure 4C). Consistent with the in vitro results, tumor volume and weight in the pc3.1-ZNF587B group were significantly decreased (Figure 4A–D). Taken together, our results implied that ZNF587B inhibited OC morphogenesis in vivo.
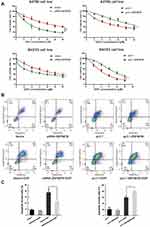
ZNF587B Enhances Cisplatin-Induced Apoptosis
CCK-8 and Annexin V+/7-AAD+ staining assays were carried out. Results of the CCK-8 assay revealed that downregulation of ZNF587B significantly increased A2780 and SKOV3 cells chemoresistance (IC50 mean values of siRNA-ZNF587B group vs vector group: 4.89 μM vs 9.52 μM in A2780; 14.37 μM vs 6.58 μM in SKOV3). The IC50 mean values were 3.17 μM and 6.57 μM in the A2780 and SKOV3 pc3.1-ZNF587B groups, which were significantly lower than those of pc3.1 groups (5.97 μM and 9.97 μM, respectively, Figure 5A). Meanwhile, the apoptotic population in the siRNA-ZNF587B group was smaller than that in the vector control group (P < 0.05, Figure 5B and C). Consistently, Annexin V+/7-AAD+ cells distinctly increased in the ZNF587B overexpression group (P < 0.05, Figure 5B and C). No apoptotic changes were observed in the groups without cisplatin. The histogram intuitively shows the percentage of apoptotic cells (Figure 5C). These data demonstrated that ZNF587B increased cisplatin-induced apoptosis in OC cells.
The Expression of ZNF587B Is Associated Prognosis
To clarify the prognostic effect of ZNF587B in OC patients, the survival data of OC patients with expression levels of ZNF587B were downloaded from the SurvExpress database (http://bioinformatica.mty.itesm.mx:8080/Biomatec/SurvivaX.jsp). In total, 82 serous cystadenocarcinoma patients were included from the ICGC dataset and were divided into low-risk and high-risk groups according to the prognostic index (Figure 6A). SurvExpress also provided the mRNA expression level of ZNF587B (Figure 6B). The results showed that the low-risk group had higher expression of ZNF587B and better prognoses (Figure 6C, HR = 1.77, 95% CI: 0.55–0.70, P = 0.023).
Discussion
Except for our previous research, almost nothing has been reported about ZNF587B.34,35 In this study, we explored the basic function of ZNF587B in OC. ZNF587B was downregulated in OC cells, while overexpression of ZNF587B in SKOV3 and A2780 cells could significantly inhibit cell proliferation, migration, and invasion. In vivo xenograft experiments also confirmed that ZNF587B could suppress tumor growth. Survival analysis showed that the overall survival of patients with low ZNF587B expression levels was dramatically shorter than that of patients with high-expression levels of ZNF587B. Taken together, we have found a novel gene that could serve as a potential tumor suppressor and prognostic biomarker. However, the mechanism by which ZNF587B affects OC remains unclear.
ZFPs are the largest transcription factor family in mammals. Approximately one-third of ZFPs are KRAB-ZFPs, and the KRAB domain is one of the most potent transcriptional repressors.21,36 KRAB-ZFPs have been reported to be involved in the regulation of cell proliferation, apoptosis, and cancer. For instance, ZNF496, a KRAB-ZFP family member, binding to estrogen receptor alpha (ERα) via its C2H2 domain, repressed the transactivation of ERα, and selectively suppressed ERα target genes, which ultimately inhibited the growth of ERα-positive breast cancer cells.37 In esophageal squamous cell carcinoma, ZNF382 suppressed Wnt/β-catenin signaling and downstream target gene expression, likely through binding directly to FZD1 and DVL2, and ultimately suppressed tumor cell proliferation and metastasis.38 ZFP82 suppresses the growth and invasion of esophageal squamous cell carcinoma cells through inhibiting NF-κB transcription.39 However, the mechanism by which ZNF587B suppresses OC cell proliferation is unknown, but likely involves the function of the KRAB domain, and remains to be explored in future studies. The major limitation of our study is that the way to detect expression of ZNF587B is only by qPCR, as there is no commercial antibody to detect its protein expression level. The conclusions based on qPCR need to be further validated by other methods and researches.
In conclusion, ZNF587B could serve as a potential tumor suppressor and prognostic indicator for OC. Our study provided a new ideal and direction for the OC diagnosis and drug development.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81974512), the Hunan Natural Science Foundation (2019JJ40485), and Graduate students independently explore innovative projects of central south university, China (2018zzts905).
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics, 2018. CA Cancer J Clin. 2018;68(4):284–296. doi:10.3322/caac.21456
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
3. Li N, Qian S, Li B, Zhan X. Quantitative analysis of the human ovarian carcinoma mitochondrial phosphoproteome. Aging. 2019;11(16):6449–6468. doi:10.18632/aging.102199
4. Li N, Zhan X. Signaling pathway network alterations in human ovarian cancers identified with quantitative mitochondrial proteomics. EPMA J. 2019;10(2):153–172. doi:10.1007/s13167-019-00170-5
5. Vaughan S, Coward JI, Bast RC
6. Christie EL, Bowtell DDL. Acquired chemotherapy resistance in ovarian cancer. Ann Oncol. 2017;28(suppl_8):viii13–viii15. doi:10.1093/annonc/mdx446
7. Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi:10.1038/sj.onc.1206933
8. Li T, Peng J, Zeng F, et al. Association between polymorphisms in CTR1, CTR2, ATP7A, and ATP7B and platinum resistance in epithelial ovarian cancer. Int J Clin Pharmacol Ther. 2017;55(10):774–780.
9. Zhai XH, Huang J, Wu FX, Zhu DY, Wang AC. Impact of XRCC1, GSTP1, and GSTM1 polymorphisms on the survival of ovarian carcinoma patients treated with chemotherapy. Oncol Res Treat. 2016;39(7–8):440–446.
10. Moxley KM, Benbrook DM, Queimado L, et al. The role of single nucleotide polymorphisms of the ERCC1 and MMS19 genes in predicting platinum-sensitivity, progression-free and overall survival in advanced epithelial ovarian cancer. Gynecol Oncol. 2013;130(2):377–382. doi:10.1016/j.ygyno.2013.04.054
11. Ali R, Alabdullah M, Miligy I, et al. ATM regulated PTEN degradation Is XIAP E3 ubiquitin ligase mediated in p85alpha deficient cancer cells and influence platinum sensitivity. Cells. 2019;8(10):1271. doi:10.3390/cells8101271
12. Fang C, Chen YX, Wu NY, et al. MiR-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (NSCLC) cells by activating the eIF3a-mediated NER signaling pathway. Sci Rep. 2017;7(1):40384. doi:10.1038/srep40384
13. Johnson SW, Swiggard PA, Handel LM, et al. Relationship between platinum-DNA adduct formation and removal and cisplatin cytotoxicity in cisplatin-sensitive and -resistant human ovarian cancer cells. Cancer Res. 1994;54(22):5911–5916.
14. Johnson SW, Laub PB, Beesley JS, Ozols RF, Hamilton TC. Increased platinum-DNA damage tolerance is associated with cisplatin resistance and cross-resistance to various chemotherapeutic agents in unrelated human ovarian cancer cell lines. Cancer Res. 1997;57(5):850–856.
15. Godwin AK, Meister A, O’Dwyer PJ, Huang CS, Hamilton TC, Anderson ME. High resistance to cisplatin in human ovarian cancer cell lines is associated with marked increase of glutathione synthesis. Proc Natl Acad Sci U S A. 1992;89(7):3070–3074. doi:10.1073/pnas.89.7.3070
16. Zhu B, Ren C, Du K, et al. Olean-28,13b-olide 2 plays a role in cisplatin-mediated apoptosis and reverses cisplatin resistance in human lung cancer through multiple signaling pathways. Biochem Pharmacol. 2019;170:113642. doi:10.1016/j.bcp.2019.113642
17. Sahel DK, Mittal A, Chitkara D. CRISPR/cas system for genome editing: progress and prospects as a therapeutic tool. J Pharmacol Exp Ther. 2019;370(3):725–735. doi:10.1124/jpet.119.257287
18. Yu JSL, Yusa K. Genome-wide CRISPR-Cas9 screening in mammalian cells. Methods. 2019;164–165:29–35. doi:10.1016/j.ymeth.2019.04.015
19. Schmierer B, Botla SK, Zhang J, Turunen M, Kivioja T, Taipale J. CRISPR/Cas9 screening using unique molecular identifiers. Mol Syst Biol. 2017;13(10):945. doi:10.15252/msb.20177834
20. Ouyang Q, Liu Y, Tan J, et al. Loss of ZNF587B and SULF1 contributed to cisplatin resistance in ovarian cancer cell lines based on genome-scale CRISPR/Cas9 screening. Am J Cancer Res. 2019;9(5):988–998.
21. Urrutia R. KRAB-containing zinc-finger repressor proteins. Genome Biol. 2003;4(10):231. doi:10.1186/gb-2003-4-10-231
22. Witzgall R, O’Leary E, Leaf A, Onaldi D, JV B. The Krüppel-associated box-A (KRAB-A) domain of zinc finger proteins mediates transcriptional repression. Proc Natl Acad Sci U S A. 1994;91(10):4514–4518. doi:10.1073/pnas.91.10.4514
23. Cassandri M, Smirnov A, Novelli F, et al. Zinc-finger proteins in health and disease. Cell Death Discov. 2017;3(1):17071. doi:10.1038/cddiscovery.2017.71
24. Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23(1):53. doi:10.1186/s12929-016-0269-9
25. Razin SV, Borunova VV, Maksimenko OG, Kantidze OL. Cys2His2 zinc finger protein family: classification, functions, and major members. Biochemistry (Mosc). 2012;77(3):217–226. doi:10.1134/S0006297912030017
26. Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;9(8):597–604. doi:10.1096/fasebj.9.8.7768350
27. Huang M, Chen Y, Han D, Lei Z, Chu X. Role of the zinc finger and SCAN domain-containing transcription factors in cancer. Am J Cancer Res. 2019;9(5):816–836.
28. Zhang Z, Chen H, Lu Y, Feng T, Sun W. LncRNA BC032020 suppresses the survival of human pancreatic ductal adenocarcinoma cells by targeting ZNF451. Int J Oncol. 2018;52(4):1224–1234.
29. Friedman JR, Fredericks WJ, Jensen DE, et al. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10(16):2067–2078. doi:10.1101/gad.10.16.2067
30. Lupo A, Cesaro E, Montano G, Zurlo D, Izzo P, Costanzo P. KRAB-zinc finger proteins: a repressor family displaying multiple biological functions. Curr Genomics. 2013;14(4):268–278. doi:10.2174/13892029113149990002
31. Wei LL, Wu XJ, Gong CC, Pei DS. Egr-1 suppresses breast cancer cells proliferation by arresting cell cycle progression via down-regulating CyclinDs. Int J Clin Exp Pathol. 2017;10(10):10212–10222.
32. Zhang H, Chen X, Wang J, et al. EGR1 decreases the malignancy of human non-small cell lung carcinoma by regulating KRT18 expression. Sci Rep. 2014;4(1):5416. doi:10.1038/srep05416
33. Zhu Z, Lou C, Zheng Z, et al. ZFP403, a novel tumor suppressor, inhibits the proliferation and metastasis in ovarian cancer. Gynecol Oncol. 2017;147(2):418–425. doi:10.1016/j.ygyno.2017.08.025
34. Lee SH, Kim HP, Kang JK, Song SH, Han SW, Kim TY. Identification of diverse adenosine-to-inosine RNA editing subtypes in colorectal cancer. Cancer Res Treat. 2017;49(4):1077–1087. doi:10.4143/crt.2016.301
35. Pelleri MC, Piovesan A, Caracausi M, Berardi AC, Vitale L, Strippoli P. Integrated differential transcriptome maps of Acute Megakaryoblastic Leukemia (AMKL) in children with or without Down Syndrome (DS). BMC Med Genomics. 2014;7(1):63. doi:10.1186/s12920-014-0063-z
36. Huntley S, Baggott DM, Hamilton AT, et al. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res. 2006;16(5):669–677.
37. Wang J, Zhang X, Ling J, et al. KRAB-containing zinc finger protein ZNF496 inhibits breast cancer cell proliferation by selectively repressing ERalpha activity. Biochimica Et Biophysica Acta Gene Regulatory Mechanisms. 2018;1861:841–853.
38. Zhang C, Xiang T, Li S, et al. The novel 19q13 KRAB zinc-finger tumour suppressor ZNF382 is frequently methylated in oesophageal squamous cell carcinoma and antagonises Wnt/β-catenin signalling. Cell Death Dis. 2018;9(5):573. doi:10.1038/s41419-018-0604-z
39. Ye L, Xiang T, Fan Y, et al. The 19q13 KRAB Zinc-finger protein ZFP82 suppresses the growth and invasion of esophageal carcinoma cells through inhibiting NF-kappaB transcription and inducing apoptosis. Epigenomics. 2019;11(1):65–80. doi:10.2217/epi-2018-0092
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.