Back to Journals » Cancer Management and Research » Volume 11
The novel lncRNA p4516 acts as a prognostic biomarker promoting gastric cancer cell proliferation and metastasis
Authors Nie ML , Han J, Huang HC, Guo T, Huangfu LT, Cheng XJ , Li XM, Du H, Li QD, Wen XZ, Ji JF
Received 16 January 2019
Accepted for publication 5 April 2019
Published 12 June 2019 Volume 2019:11 Pages 5375—5391
DOI https://doi.org/10.2147/CMAR.S201793
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Kenan Onel
Meng-Lin Nie,1 Jing Han,1 Han-Chen Huang,2,3 Ting Guo,1 Long-Tao Huangfu,1 Xiao-Jing Cheng,1 Xiao-Mei Li,1 Hong Du,1 Qing-Da Li,1 Xian-Zi Wen,1 Jia-Fu Ji1,4
1Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Division of Gastrointestinal Cancer Translational Research Laboratory, Peking University Cancer Hospital & Institute, Beijing, People’s Republic of China; 2Key Laboratory of RNA Biology, Beijing Key Laboratory of Noncoding RNA, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, People’s Republic of China; 3Centre for Cognitive Machines and Computational Health (CMaCH), The School of Electronic Information and Electrical Engineering, Shanghai Jiao Tong University, Shanghai, People’s Republic of China; 4Department of Gastrointestinal Surgery, Peking University Cancer Hospital, Beijing, People’s Republic of China
Purpose: Emerging evidence has shown that long noncoding RNAs (lncRNAs) participate in oncogenesis and tumor progression. We previously found a novel lncRNA p4516 which was closely associated with prognosis by preliminary study of lncRNA expression profile from paired tumors and nontumor tissues in 198 gastric cancer (GC) patients. However, the exact biological functions and the underlying molecular mechanisms of p4516 in gastric tumorigenesis still remain unclear.
Materials and methods: The RNA fluorescence in situ hybridization (RNA-FISH) analysis, cytoplasmic and nuclear RNA isolation and qRT-PCR were applied to determine the subcellular localization of p4516. Expression levels of p4516 were assessed using qRT-PCR in both GC cell lines and in 142 primary GC tissues. Correlations between p4516 expression and GC patients’ clinicopathological parameters were analyzed. Gain- and loss-of-function experiments were employed to investigate the role of p4516 in proliferation, migration and invasion both in vitro and in vivo. In addition, Western blotting and immunohistochemical staining were used to examine the protein expression levels.
Results: LncRNA p4516 was mainly localized in the nucleus of GC cells and p4516 tended to have higher expression levels in GC cells compared to the normal gastric mucosa-derived cells GES-1. Furthermore, higher expression levels of p4516 correlated with worse clinical outcomes in GC patients and acted as an independent prognostic biomarker. Functional analysis revealed that p4516 participated in the regulation of GC cell proliferation, invasion and migration both in vivo and in vitro. Moreover, p4516 was involved in epithelial–mesenchymal transition (EMT) in GC cells.
Conclusion: Our study demonstrated the oncogenic role of novel lncRNA p4516 in the gastric carcinogenesis for the first time. High expression of p4516 may act as prognostic marker in patient with gastric cancer.
Keywords: LncRNA, p4516, Gastric cancer, EMT
Introduction
Gastric cancer (GC) is the second leading cause of cancer-related death worldwide1 and the incidence of GC has a distinct geographical distribution.2 Early detection can largely improve the 5-year survival rate;3 however, due to lack of effective diagnostic and therapeutic strategies, more than 80% GC patients in China had developed to an advanced stage at the time of diagnosis and the 5-year survival rate is less than 20%.4 Therefore, to identify novel biomarkers for the diagnosis and prognosis of GC as well as to develop the therapeutic targets are needed.
Long non-coding RNAs (lncRNAs) are a class of pervasively transcribed products more than 200 nt with no obvious coding capacities.5 They are initially considered to be useless of genomic transcriptions.6 However, increasing evidence gradually revealed the numerous gene regulatory functions of these so-called “junk gene”, involved in imprinting,7 epigenetic regulation,8 transcriptional and post-transcription regulation. They contributed to many physiological and pathological processes including oncogenesis and cancer progression.9 Aberrant expression of several lncRNAs are observed in GC cells. For example, HOTAIR, UCA1, H19 and GHET1 facilitate the proliferation and metastasis procedures of GC,10,11,12,13 whereas MEG3, MT1JP and ADAMTS9-AS2 act in the opposite way.14,15,16 LncRNAs also have significant clinical implications in GC patients.17 A higher expression level of HOTAIR was more prone to exit in the diffuse types of GC and found to be correlated with a shorter overall survival (OS) in the diffuse type of GC patients.18 Correlation analysis between lncRNA CASC11 expression and clinical pathological factors revealed that the high expression of CASC11 was significantly associated with larger tumor size, lymph node metastasis and TNM staging in GC patients.19
The roles of epithelial–mesenchymal transition (EMT) and the reverse process, mesenchymal-to-epithelial transition (MET), were initially studied in the context of embryonic morphogenesis.20,21 EMT process plays a central part in various pathological processes, including wound healing, tissue fibrosis and carcinoma progression.22,23 The survival of patients with local infiltration, lymph node or distant organ metastasis is unfavorable; therefore, the investigation of the mechanisms regarding EMT in GC has become a major focus.24 Many different lncRNAs have been found to be involved in the EMT process. For instance, the lncRNA SNHG6 promoted EMT through epigenetic silencing of p27 and sponging miR-101-3p in GC,25 and lncRNA UCA1 augmented the invasive and migratory capabilities of GC cells by regulating the expression of EMT-associated factors.26
Previously, we conducted a gene expression profiling on 198 GC and paired adjacent nontumor tissues from Chinese patients by lncRNA array with 40,914 transcripts. Then, a recently proposed analytical method namely FDA-based PT-test was performed according to the Table 3 & 4 from the previous work by Xu27, which was designed to screen prognostic marker of lncRNAs based on gene expression profiling data of tumor and paired nontumor tissues. As a result, a set of lncRNAs, including a novel lncRNA p4516 (ID: ENST00000563647.1, refer to Ensembl genome browser 94), closely associating with prognosis were revealed (unpublished data). p4516, as a novel lncRNA, resides on chromosome 14 with 1 exon and is 981 base pairs in length without coding protein. In the present study, we demonstrated for the first time that the completely novel lncRNA p4516 was not only associated with gastric carcinogenesis but also served as a poor prognostic marker in GC patients, thus provided a novel potential target for gastric cancer treatment.
Materials and methods
Patient samples
A total of 142 GC tissues were obtained from patients who were diagnosed and underwent radical resections at Peking University Cancer Hospital between 2007 and 2010 and followed up to March 2016. All the patients signed informed consent forms, and this was conducted in accordance with the Declaration of Helsinki. Tumor stages were classified based on the 2010 tumor node metastasis (TNM) classification recommended by the American Joint Committee on Cancer (AJCC 7th edition). T and N classifications were evaluated according to the final pathological result, whereas the M classification was determined by surgical and imaging findings. None of the patients received local or systemic treatment prior to surgery. All the resected tissues were immediately frozen in liquid nitrogen, stored at −80°C and prepared for RNA extraction use. Clinicopathological parameters and follow-up information were collected from available patient data. This study was approved by Ethics Committee of Peking University Cancer Hospital (Approval number: 2018KT07).
Cell culture
The GC-derived cell lines: BGC823, HGC27, MGC803, N87 and SGC7901, and the normal gastric mucosa-derived cell line GES-1 were obtained from Chinese National Infrastructure of Cell Line Resource. All the cell lines were cultured in Dulbecco’s modified Eagle medium (DMEM; GIBCO, Carlsbad, NY, USA) supplemented with 10% fetal bovine serum (FBS; GIBCO, Carlsbad, NY, USA), and maintained in a 37°C incubator with a 5% CO2.
RNA extraction and qRT-PCR
Total RNA was extracted from tissue samples and cell lines using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. First-strand cDNA was generated by reverse transcription polymerase chain reaction using a reverse transcription system kit (Invitrogen, Carlsbad, CA, USA). Quantitative real-time PCR (qRT-PCR) was performed with the ABI PRISM 7500 Sequence Detection System according to the SYBR Green method (amplification condition for p4516: 95℃ 10 min followed by 45 cycles of 95℃ 15 s and 60°C 1 min). For each sample, gene expression was normalized to GAPDH or U6. The primer sequences used in this research were listed in Table S1. The qRT-PCR reactions for each sample were performed in triplicate, and the relative RNA expression level was calculated using the comparative Ct method. Cutoff Finder (http://molpath.charite.de/cutoff/) is an easy-to-use web application for the determination of cutoff points in molecular data.28 We used the Cutoff Finder to calculate the cutoff point of the p4516 expression levels according to 2−ΔCt value (cutoff point =0.0022, ΔCt=Ct target-Ct reference).
Establishment of p4516 knockdown or overexpression cell lines
Knocking-down of p4516 procedure was carried out as described previously.29 Lentivirus was produced by the co-transfection of 293T cells with a pLenti vector (psi-LVRH1GP -shControl or psi-LVRH1GP -shp4516) and lentiviral packaging mix (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. shRNAs against lncRNA p4516 were designed with MIT’s siRNA designer (http://sirna.wi.mit.edu/home.php). At least four quadruplexes were designed, and the most effective shRNAs were used for subsequent studies (Figure S1). The effective targeting sequence against p4516 is as below: 5ʹ-GCAGATAATGCATCTTCCAAG-3ʹ. Lentivirus-containing supernatant was harvest at 48 hrs post-transfection, purified by centrifugation and stored at −80°C. For viral transductions, 1 mL of the scrambled shControl or shp4516 lentiviruses were incubated with BGC823 cells overnight at 37°C in the cell culture incubator. BGC823 cells with stable expression of depleted endogenous p4516 were selected with puromycin (0.8 μg/mL) in the culture media.
Full length of p4516 (ENST00000563647.1) was synthesized with the following primers: FP: 5ʹ-CCCaagcttTGTCATAGAAACTATATAGCATTGA-3ʹ; RP: 5ʹ-CGggatccTTGCATTTTAATAAACTTAATTT-3, and sub-cloned into the pcDNA3.1 (+) vector (Viewsolid biotech, Beijing, China) to generate pcDNA3.1 (+)-p4516 vector. SGC7901 cells were seeded in six-well plates at 70% confluence before transfection. Transfections were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. After 48 hrs post-transfection, G418 was added at a concentration of 800 μg/mL to establish stable cell line of SGC7901 with p4516 overexpression.
RNA fluorescence in situ hybridization
The RNA fluorescence in situ hybridization (RNA-FISH) analysis was used to detect the subcellular location of lncRNA p4516 using a digoxin-labeled RNA probe (Table S2), which was designed and synthesized according to the whole sequence of p4516. BGC823 and SGC7901 cells were fixed in 4% formaldehyde for 15 mins and then washed with PBS. The fixed cells were treated with pepsin (1% in 10 mM HCl) and dehydrated in a gradient series of increasing ethanol (70%, 90% and 100%). The cells were then dried in air and further incubated with 40 nM of the RNA probe-p4516 in hybridization buffer (100 mg/mL dextran sulfate, 10% formamide in 2× saline sodium citrate) at 80°C for 2 mins. Hybridization was conducted at 55°C for 2 hrs before the slides were washed and dehydrated. The air-dried slides were then mounted with Prolong Gold Antifade Reagent with 40,6-diamidino-2-phenylindole (DAPI) was used for nuclear detection. Images were captured using LSM 800 confocal microscope (Carl Zeiss, Jena, Germany). Nuclei were labeled with DAPI (blue) and p4516 were labeled by the RNA probe-p4516 (red).
Subcellular fractionation
For nuclear and cytoplasmic fraction separation, RNAs from BGC823 and SGC7901 cells were isolated by using the PARIS kit (Life Technologies, USA) according to the manufacturer’s instructions. The RNA expression level of p4516 in nuclear and cytoplasmic fractions was determined by qRT-PCR. U6 was a positive control for nuclear fraction and GAPDH was a positive control for cytoplasmic fraction. The primer sequences are listed in Table S1.
Cell proliferation assay
Cells were seeded in 96-well plates at a density of 3×103/well. Cell proliferation was monitored with IncuCyte live cell analysis imaging system (Essen Bioscience, Ann Arbor, MI, USA).
Colony formation assay
For the colony formation assay, the established stable cell lines were seeded into 6-well plates at 600 cells/well and incubated at 37°C with 5% CO2 for 14 days. The cells were then washed twice with PBS carefully and fixed with 75% ethyl alcohol for 15 mins at room temperature. The cells were stained with 0.1% crystal violet, and the colonies containing ≥50 cells were counted.
Migration and invasion assay
Transwell chambers, with or without Matrigel (Corning Life Science, Corning, NY, USA), were used to examine the cell migration and invasion, respectively. A total of 3×104 cells were suspended in 200 μL serum-free DMEM and seeded onto polycarbonate filters, which had been either precoated with 100 μL of Matrigel for the invasion assay or left uncoated for the migration assay. A total of 600 μL of DMEM containing 10% FBS was loaded into the lower chamber.
Wound-healing assay
Cell motility was assessed by wound-healing assay. Briefly, wounds were generated by using a 200-μL plastic pipette tip to scratch when cells reached 100% confluence. Growth medium was replaced with serum-free medium, and the wound gaps were monitored with real-time IncuCyte live cell analysis system.
Western blot assays
Proteins were extracted by a pre-chilled RIPA lysis buffer (Solarbio, CN) containing protease and phosphatase inhibitor cocktail (Roche, Basel, Switzerland) and quantified using a bicinchoninic acid (BCA) protein quantification kit (Thermo, USA). Total protein lysates were separated by SDS-PAGE and then transferred onto polyvinylidene difluoride (PVDF) membranes (Whatman, Germany). The PVDF membranes were blocked with 1% bull serum albumin and incubated with specific primary antibodies at 4°C overnight followed by the secondary antibody incubation at room temperature for 1 hr. Antibody against E-cadherin, Vimentin, Snail and ZEB1 were purchased from Cell Signaling Technology (#9782). All primary antibodies were diluted at 1:1000, and the corresponding secondary antibodies were diluted at 1:5000.
In vivo xenograft mouse model
Female athymic BALB/c nude mice (4–6 weeks, 18–20 g) were used as host mice. The animals were bred in a specific pathogen-free environment at the Laboratory Animal Center of the Peking University Cancer Hospital and given sterilized food and tap water ad libitum.
Animal studies were carried out in strict adherence with institutional guidelines, and they were approved by the Animal Ethics Committee at Peking University Cancer Hospital (Number: EAEC 2018–22). BGC823-shp4516 cells and BGC823-shControl cells (approximately 5×105 cells/200 μL per mouse) were subcutaneously injected into the left or right subaxillary, respectively. Caliper was used to measure the width (W) and length (L) of the xenograft and the volumes of xenograft was calculated by the following formula: V=0.5×L×W2. An electronic balance (Sartorius, BSA224S-CW, Germany) was used to measure the weight of xenografts. The mice were sacrificed 3 weeks after injection, and the xenografts were fixed with formalin and embedded with paraffin.
BGC823 cells with or without stable expression of p4516 shRNA (5×106 cells/400 μL volume per mouse) were injected into the BALB/c nude mice (n=5 per group) via tail vein. Four weeks after injection, all the mice were sacrificed and the lungs were collected. Bouin’s solution was injected from the main bronchi to fix the lung tissues.
All the dissected tissues were formalin-fixed and paraffin-embedded for H&E staining and ki-67 immunohistochemical staining.
Statistical analysis
Statistical analysis was performed with the SPSS 21.0 software package (IBM Corporation, Armonk, NY, USA). Kaplan–Meier method and log-rank test were used to calculate 5-year overall survival (OS). Multivariate analysis was used to detect the independent prognostic parameters. Relationships between p4516 expression and clinicopathological characteristics were analyzed by the Chi-square tests. Two-tailed Student’s t-test was used to test the differences between groups (independent sample t-test). All experiments were repeated at least three times. Differences were defined as significant as follows: *p<0.05; **p<0.01; ***p<0.001.
Results
Identification of lncRNA p4516
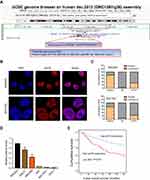
Previously, we conducted a gene expression profiling on 198 GC and paired adjacent tissues from Chinese patients by lncRNA array with 40,914 transcripts. We found that p4516 (probe name) (probe sequence: 5ʹ-GGAGAGCTACTGTACTGACTGTACTTGTGGAATGCAGCGCTTCATTAAATTAAGTTTATT-3ʹ) was closely associated with GC patient survival (data has not published yet). For the identification of p4516, BLAT search (http://genome.ucsc.edu/index.html) was done in order to infer its genomic coordinates. It is annotated as AL049838.1–201 (ENST00000563647.1) and resides on chromosome 14 in human. LncRNA p4516 is composed of one exon, with a full length of 981 bp, and the annotation on Ensembl Genome Browser 94 confirmed that it has no protein-coding potential (Figure 1A). It has been known that lncRNA function can be largely affected by its subcellular localization; accordingly, we performed RNA fluorescence in situ hybridization (RNA-FISH) to detect the p4516 subcellular localization in BGC823 cells and SGC7901 cells. We found that p4516 was mainly distributed in the nucleus (Figure 1B). This result was further verified by cytoplasmic and nuclear RNA fractionation analysis by qRT-PCR (Figure 1C).
High expression of lncRNA p4516 predicted adverse phenotypes and poor prognosis of GC patients
We next evaluated the expression levels of p4516 in GC cell lines and primary GC tissues by qRT-PCR. It revealed that p4516 was highly expressed in GC-derived tumor cell lines compared to the normal gastric mucosa-derived cell line GES-1 (Figure 1D). To clarify the relationship between the p4516 expression level and clinicopathological parameters, the GC patients (n=142) were classified according to the p4516 expression level into high-p4516 group (cutoff ≥0.0022) and low-p4516 group (cutoff <0.0022). As shown in Table 1, high expression of p4516 was positively associated with poor differentiation (P=0.011), vascular invasion (P=0.007), depth of invasion (P=0.010), lymphatic metastasis (P=0.036), distant metastasis (P=0.015) and TNM stage (P=0.017).
 | Table 1 Correlation between p4516 expression levels and clinicopathological features in patients with gastric cancer |
Furthermore, we assessed the prognostic significance of GC related parameters. Univariate analysis results showed that tumor size, histologic differentiation, vascular invasion, depth of invasion, lymphatic metastasis, distant metastasis, TNM stage and p4516 expression were used as prognostic factors for 5-year overall survival (OS) (Table S3). Multivariate analysis of 5-year overall survival (OS) indicated that high expression of p4516 was an independent marker for poor prognosis (p=0.002) (Table 2). Kaplan–Meier survival curves further demonstrated that 5-year OS was worse in GC patients with high p4516 expression than in patients with lower p4516 expression (p=0.001, Figure 1E).
 | Table 2 Multivariate analysis of prognostic parameters in patients with gastric cancer by Cox regression analysis |
Effects of lncRNA p4516 on proliferation and colony formation of GC cells in vitro
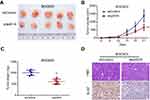
To determine the potential effects of p4516 on GC cell growth and cell mobility, we first established stable BGC823 cells with p4516 depletion and SGC7901 cell line with p4516 overexpression, respectively. qRT-PCR assay was used to determine the efficacy of transfection (Figure 2A and B). The effect of p4516 on cell proliferation was evaluated by an IncuCyte live cell analysis system. Our results showed that cell proliferation was significantly inhibited in the p4516 knockdown cells, but dramatically increased in the p4516 overexpression cells (Figure 2C and D). Similarly, the colony formation capacity was suppressed in the p4516 knockdown cells but elevated in the overexpression cells (Figure 2E and F).
Downregulation of p4516 inhibited tumorigenesis in vivo
To further determine the tumorigenic ability of p4516 on GC cell lines, xenograft tumor models were established by subcutaneous injection of BGC823 cells with or without p4516 depletion. Tumors exhibited a smaller size and slower growth rate in the BGC823-shp4516-injected group compared with the BGC823-shControl-injected group (Figure 3A-C). Immunohistochemical staining analysis showed that the ki-67 expression levels in tumor tissues derived from BGC823-shp4516 cells were significantly decreased compared with ones derived from the BGC823-shControl cells (Figure 3D). These results demonstrated that downregulation of p4516 inhibited the tumorigenic ability of GC cells in vivo.
Lncrna p4516 promoted migration and invasion of GC cell lines both in vitro and in vivo
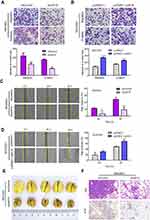
As the high expression level of p4516 was associated with vascular invasion, lymph node and distant metastasis in patients with primary GC, we assumed that p4516 may endow GC cells with invasive behavior. As we expected, Matrigel invasion and wound healing assays showed that BGC823-shp4516 cells revealed decreased migration and invasion compared to shControl cells, while SGC7901 cells with p4516 overexpression showed the opposite results (Figure 4A–4D).
To examine the effects of p4516 on tumor metastatic colonization, BGC823-shp4516 cells or BGC823-shControl cells were injected into athymic nude mice via the tail vein. Metastatic potential was assessed by counting colonized tumor nodules in the lung. BGC823-shp4516-injected mice had fewer lung tumor nodules compared to the BGC823-shControl-injected mice (Figure 4E). This difference was further confirmed with hematoxylin and eosin (H&E) and ki-67 staining of the lung tissue sections (Figure 4F). Taken together, these data indicate that p4516 aggressively promoted cell migration and invasion in GC.
LncRNA p4516 enhanced EMT in GC cells
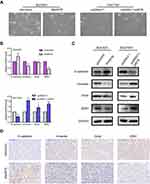
Enhanced cell migration and invasion capabilities are important consequences of EMT, an early event in tumor metastasis. Cell morphology changes were detected in p4516 depleted BGC823 cells and overexpressed SGC7901 cells. We found that p4516 depletion in BGC823 cells changed cell morphology from an elongated spindle-like appearance to a cobblestone-like appearance, whereas p4516 overexpressed SGC7901 cells exhibited an elongated fibroblast phenotype compared to the control cells (Figure 5A). Therefore, we examined EMT-associated markers by qRT-PCR, Western blotting and immunohistochemical staining. The results showed that overexpression of p4516 could decrease the expression of epithelial marker E-cadherin, and increase the expression of mesenchymal markers of Vimentin, Snail and ZEB1 both in transcript and protein level in SGC7901 cells (Figure 5B lower panel and Figure 5C right panel). In contrast, knockdown of p4516 upregulated E-cadherin and down-regulated Vimentin, Snail and ZEB1 expression both in transcript and protein levels in BGC823 cells (Figure 5B upper panel, Figure 5C left panel and Figure 5D). All these data suggested that the role of p4516 in promoting GC cells’ metastasis was partly attributed to affecting EMT process, and further experiments are still needed to elucidate the potential mechanism.
Discussion
Gastric cancer patients at advanced stages are prone to have a lower 5-year overall survival rate, which is largely due to the lymph nodes and distant organs metastasis.30,31,32 Therefore, it is seriously imperative to explore prognostic markers and underlying molecular mechanisms.
Increasing evidence has proved that lncRNAs play a vital role in carcinogenesis and metastasis of GC. LncRNA p4516 is a newly discovered non-coding RNA showing strong association with prognosis according to our preliminary study on lncRNA expression profile. However, the biological functions and molecular mechanisms of p4516 in gastric carcinogenesis have been unknown yet.
In the present study, we first analyzed the association between p4516 expression and clinicopathological parameters. LncRNA p4516 expression level was positively correlated with poor differentiation, vascular invasion, depth of invasion, lymph node and distant metastasis, and more advanced cancer stage. Furthermore, high expression of p4516 indicated poor survival of the patients with GC. This is consistent with the other studies showing that the lncRNA was associated with prognostic value in GC.33 For example, higher expression levels of TP73-AS1 and GACAT3 were predicted to have shorter overall survival in GC.34,35 Those data provided further supports that p4516 could be a potential independent biomarker for the prognosis of GC patients.
In the past decades, lncRNAs had been considered as non-functional molecules, but emerging evidence started to suggest the importance of lncRNAs function linked to the tumor progression. For explaining above observed phenomenon, gain- and loss-of-function approaches were applied. Depletion of p4516 demonstrated significant anti-growth and anti-invasiveness activity in BGC823 cells, and overexpression of p4516 led to the adverse results. Given that enhanced capabilities of cell migration and invasion are important consequence of EMT, and in light of morphological changes of GC cells caused by p4516 knockdown or overexpression, we presume that p4516 may contribute to EMT. As a result, p4516 knockdown or overexpression resulted in alterations in EMT-associated protein expression. Thus, these data together could explain our findings that high expression of p4516 was significantly correlated with metastasis and poor survival in patients with GC.
Metastasis is one of the typical hallmarks of cancer, and it is also the most lethal cause in cancer patients, therefore focusing on tumor cell metastasis regulators could be the potential targets to therapeutic application.36 A critical event in driving stationary tumor cells to disseminate from the primary tumors and migrate to remote organs is the EMT process.23 EMT is a complex process involved in cancer metastasis and is characterized by increasing migratory potential of epithelial cells and transforming those cells to the invasive mesenchymal cells.37 Existing evidence proved that lncRNA could modulate cancer metastasis via affecting EMT in GC.16,38,39,40 In this study, both knockdown and overexpression of p4516 altered the expression of EMT-associated proteins, indicating the oncogenic role of p4516 in GC, at least partially via regulating EMT process.
LncRNA can regulate gene expression by distinct molecular mechanisms, mainly at epigenetic, transcription and post-transcription level.41,42 Subcellular localization of lncRNA often gives us some clues for exploring the underlying molecular mechanism of their roles. Generally, lncRNA in the cytoplasm functions in a post-transcriptional manner, whereas that within nucleus acts on transcriptional and/or epigenetic regulation by interacting with chromatin remodeling complexes.43,44 Our observation of p4516 expression is mainly localized in the cell nucleus of GC cells, suggesting that the oncogenic roles of p4516 in GC may through chromatin modification or/and transcriptional regulation. Further experimental assays, such as RNA pull-down, mass spectrometry, RNA binding protein immunoprecipitation and RNA sequencing should be applied for clarifying the underlying molecular mechanism.
Conclusion
In summary, we report the potential role of p4516 for the first time. In the present study, high expression of p4516 was associated with poor prognosis of patients with GC. In addition, p4516 enhanced GC cell proliferation and invasiveness by promoting EMT, indicating that p4516 plays an important role during gastric tumorigenesis, and it is expected to act as a potential prognostic biomarker in GC.
Acknowledgments
This work was supported by the Natural Science Foundation of Beijing (No. 7132051), the National Natural Science Foundation of China (Nos. 81301874, 81802471), the Interdisciplinary medicine Seed Fund of Peking University (No. BMU2018MX018), the General Program Research Fund for the Doctoral Program of Higher Education (No. 2012001120135), the Science Foundation of Peking University Cancer Hospital (2017-23, A001538), clinical medicine + X special fund from Peking University and Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding Support (No. ZYLX201701). Han-Chen Huang was partly supported by the Zhi-Yuan chairprofessorship start-up grant (WF220103010) from Shanghai Jiao Tong University.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
3. Wadhwa R, Taketa T, Sudo K, Blum MA, Ajani JA. Modern oncological approaches to gastric adenocarcinoma. Gastroenterol Clin North Am. 2013;42(2):359–369. doi:10.1016/j.gtc.2013.01.011
4. Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388(10060):2606. doi:10.1016/S0140-6736(16)32226-7
5. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10(3):155–159. doi:10.1038/nrg2521
6. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
7. Lee JT, Bartolomei MS. X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell. 2013;152(6):1308–1323. doi:10.1016/j.cell.2013.02.016
8. Shen C, Zhong N. Long non-coding RNAs: the epigenetic regulators involved in the pathogenesis of reproductive disorder. Am J Reprod Immunol. 2015;73(2):95–108. doi:10.1111/aji.12315
9. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi:10.1038/nm.3981
10. Feng X, Huang S. Effect and mechanism of lncRNA HOTAIR on occurrence and development of gastric cancer. J Cell Biochem. 2017;120:6899-6907. doi:10.1002/jcb.26594.
11. Wang ZQ, He CY, Hu L, et al. Long noncoding RNA UCA1 promotes tumour metastasis by inducing GRK2 degradation in gastric cancer. Cancer Lett. 2017;408:10–21. doi:10.1016/j.canlet.2017.08.013
12. Chen JS, Wang YF, Zhang XQ, et al. H19 serves as a diagnostic biomarker and up-regulation of H19 expression contributes to poor prognosis in patients with gastric cancer. Neoplasma. 2016;63(2):223–230. doi:10.4149/207_150821N454
13. Huang H, Liao W, Zhu X, Liu H, Cai L. Knockdown of long noncoding RNA GHET1 inhibits cell activation of gastric cancer. Biomed Pharmacother. 2017;92:562–568. doi:10.1016/j.biopha.2017.05.088
14. Dan J, Wang J, Wang Y, et al. LncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed Pharmacother. 2018;99:931–938. doi:10.1016/j.biopha.2018.01.164
15. Xu Y, Zhang G, Zou C, et al. LncRNA MT1JP suppresses gastric cancer cell proliferation and migration through MT1JP/MiR-214-3p/RUNX3 axis. Cell Physiol Biochem. 2018;46(6):2445–2459. doi:10.1159/000489651
16. Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed Pharmacother. 2018;107:185–193. doi:10.1016/j.biopha.2018.06.146
17. Gu Y, Chen T, Li G, et al. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol. 2015;11(17):2427–2441. doi:10.2217/fon.15.175
18. Endo H, Shiroki T, Nakagawa T, et al. Enhanced expression of long non-coding RNA HOTAIR is associated with the development of gastric cancer. PLoS One. 2013;8(10):e77070. doi:10.1371/journal.pone.0077070
19. Zhang L, Kang W, Lu X, Ma S, Dong L, Zou B. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17(15):1886–1900. doi:10.1080/15384101.2018.1502574
20. Shook D, Keller R. Mechanisms, mechanics and function of epithelial-mesenchymal transitions in early development. Mech Dev. 2003;120(11):1351–1383.
21. Hay ED. The mesenchymal cell, its role in the embryo, and the remarkable signaling mechanisms that create it. Dev Dyn. 2005;233(3):706–720. doi:10.1002/dvdy.20345
22. Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119(6):1420–1428. doi:10.1172/JCI39104
23. Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. doi:10.1016/j.cell.2016.06.028
24. Peng Z, Wang CX, Fang EH, Wang GB, Tong Q. Role of epithelial-mesenchymal transition in gastric cancer initiation and progression. World J Gastroenterol. 2014;20(18):5403–5410. doi:10.3748/wjg.v20.i18.5403
25. Yan K, Tian J, Shi W, Xia H, Zhu Y. LncRNA SNHG6 is associated with poor prognosis of gastric cancer and promotes cell proliferation and EMT through epigenetically silencing p27 and sponging miR-101-3p. Cell Physiol Biochem. 2017;42(3):999–1012. doi:10.1159/000478682
26. Zuo ZK, Gong Y, Chen XH, et al. TGFbeta1-induced LncRNA UCA1 upregulation promotes gastric cancer invasion and migration. DNA Cell Biol. 2017;36(2):159–167. doi:10.1089/dna.2016.3553
27. Xu L. A new multivariate test formulation: theory, implementation, and applications to genome-scale sequencing and expression. Appl Inf. 2016;3(1). doi:10.1186/s40535-015-0016-4
28. Budczies J, Klauschen F, Sinn BV, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. doi:10.1371/journal.pone.0051862
29. Li S, Li Z, Guo T, et al. Maternal embryonic leucine zipper kinase serves as a poor prognosis marker and therapeutic target in gastric cancer. Oncotarget. 2016;7(5):6266–6280. doi:10.18632/oncotarget.6673
30. Digklia A, Wagner AD. Advanced gastric cancer: current treatment landscape and future perspectives. World J Gastroenterol. 2016;22(8):2403–2414. doi:10.3748/wjg.v22.i8.2403
31. Yamashita K, Ema A, Hosoda K, et al. Macroscopic appearance of Type IV and giant Type III is a high risk for a poor prognosis in pathological stage II/III advanced gastric cancer with postoperative adjuvant chemotherapy. World J Gastrointest Oncol. 2017;9(4):166–175. doi:10.4251/wjgo.v9.i4.166
32. Machlowska J, Maciejewski R, Sitarz R. The pattern of signatures in gastric cancer prognosis. Int J Mol Sci. 2018;19(6). doi:10.3390/ijms19061658
33. Li F, Huang C, Li Q, Wu X. Construction and comprehensive analysis for dysregulated long non-coding RNA (lncRNA)-associated competing endogenous RNA (ceRNA) network in gastric cancer. Med Sci Monit. 2018;24:37–49.
34. Wang Y, Xiao S, Wang B, Li Y, Chen Q. Knockdown of lncRNA TP73-AS1 inhibits gastric cancer cell proliferation and invasion via the WNT/beta-catenin signaling pathway. Oncol Lett. 2018;16(3):3248–3254. doi:10.3892/ol.2018.9040
35. Feng L, Zhu Y, Zhang Y, Rao M. LncRNA GACAT3 promotes gastric cancer progression by negatively regulating miR-497 expression. Biomed Pharmacother. 2018;97:136–142. doi:10.1016/j.biopha.2017.10.074
36. Fouad YA, Aanei C. Revisiting the hallmarks of cancer. Am J Cancer Res. 2017;7(5):1016–1036.
37. O‘Leary K, Shia A, Schmid P. Epigenetic regulation of EMT in non-small cell lung cancer. Curr Cancer Drug Targets. 2018;18(1):89–96. doi:10.2174/1568009617666170203162556
38. Qi P, Lin WR, Zhang M, et al. E2F1 induces LSINCT5 transcriptional activity and promotes gastric cancer progression by affecting the epithelial-mesenchymal transition. Cancer Manag Res. 2018;10:2563–2571. doi:10.2147/CMAR.S171652
39. Weng J, Xiao J, Mi Y, et al. PCDHGA9 acts as a tumor suppressor to induce tumor cell apoptosis and autophagy and inhibit the EMT process in human gastric cancer. Cell Death Dis. 2018;9(2):27. doi:10.1038/s41419-018-1111-y
40. Xu W, He L, Li Y, Tan Y, Zhang F, Xu H. Silencing of lncRNA ZFAS1 inhibits malignancies by blocking Wnt/beta-catenin signaling in gastric cancer cells. Biosci Biotechnol Biochem. 2018;82(3):456–465. doi:10.1080/09168451.2018.1431518
41. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi:10.1016/j.bbagrm.2014.08.012
42. Fatima R, Akhade VS, Pal D, Rao SM. Long noncoding RNAs in development and cancer: potential biomarkers and therapeutic targets. Mol Cell Ther. 2015;3:5. doi:10.1186/s40591-015-0042-6
43. Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41(9):761–772. doi:10.1016/j.tibs.2016.07.003
44. Zhang K, Shi ZM, Chang YN, Hu ZM, Qi HX, Hong W. The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene. 2014;547(1):1–9. doi:10.1016/j.gene.2014.06.043
Supplementary material
 | Figure S1 Four different shRNA knocking-down efficiency. |
 | Table S1 Primers used in this research |
 | Table S2 RNA probe sequence used in FISH analysis |
 | Table S3 Univariate analysis of prognostic parameters in patients with gastric cancer |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.