Back to Journals » Cancer Management and Research » Volume 10
The mutation of hepatitis B virus and the prognosis of hepatocellular carcinoma after surgery: a pilot study
Authors Zhang Y , Huang J , Chen J , Yang K, Chen J, Xu L , Zhou Z, Chen M
Received 16 December 2017
Accepted for publication 29 January 2018
Published 27 March 2018 Volume 2018:10 Pages 599—611
DOI https://doi.org/10.2147/CMAR.S160047
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Yaojun Zhang,1,2,* Junting Huang,1,2,* Jinbin Chen,1,2 Keli Yang,1,2 Jiancong Chen,1,2 Li Xu,1,2 Zhongguo Zhou,1,2 Minshan Chen1,2
1Department of Hepatobiliary Surgery, Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China; 2State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Background: Although hepatitis B virus (HBV) is still one of the most common etiological factors for hepatocellular carcinoma (HCC), the association between the HBV mutations and the clinical characteristics and prognosis of HBV-related HCC patients (HBV-HCC) after surgical resection remains largely unknown.
Materials and methods: A cohort of 131 consecutive patients who received hepatectomy for HBV-HCC were retrospectively enrolled. The HBV genotype and 14 genomic mutations, which have been reported to relate to HCC in liver samples, were sequenced. The associations between the genomic mutations and clinical characteristics and outcomes were analyzed.
Results: Both A1762T/G1764A mutation and Pre S deletion related to worse overall survival (OS, p=0.040 and p<0.001, respectively) and disease-free survival (DFS, p=0.040 and p<0.001, respectively), G1899A mutation related to worse OS (p=0.030), A1762T/G1764A mutation correlated with tumor size (r=0.204, p=0.019), G1899A mutation correlated with vascular invasion (r=0.332, p<0.001), and Pre S deletion correlated with alpha-fetoprotein (AFP; r=0.254, p=0.003) positively. Multivariate analysis with Cox proportional hazards model revealed that both A1762T/G1764A mutation and Pre S deletion were independent prognostic factors for OS (hazard ratio [HR]=3.701, 95% CI=1.390–9.855, p=0.009, and HR=4.816, 95% CI=2.311–10.032, p<0.001, respectively) and DFS (HR=3.245, 95% CI=1.400–7.521, p=0.006, and HR=2.437, 95% CI=1.311–4.530, p<0.001, respectively), and patients with dual mutations were found to have the worst OS and DFS (p<0.001 and p<0.001, respectively). Patients with A1762T/G1764A mutation or Pre S deletion were more likely to have early recurrence (p=0.042 and p=0.019, respectively).
Conclusion: HBV DNA genomic mutations in A1762T/G1764A and Pre S deletion were associated with worse prognoses and early recurrence for HBV-HCC patients after surgery.
Keywords: hepatitis B virus, mutation, hepatocellular carcinoma, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer mortality worldwide. Approximately 700,000 people die of HCC each year.1,2 The prognosis of HCC is poor because the disease is often at a fairly advanced stage at the time of diagnosis.3,4 Liver resection remains the main curative option for HCC with a 5-year overall survival (OS) rate of 50–70% after curative hepatectomy. However, the long-term prognosis is still unsatisfactory due to the high recurrence rate.3,5 To improve prognosis, it is crucial to identify the risk factors affecting hepatocarcinogenesis and post-surgical recurrence of HCC.
Despite successful vaccination programs, hepatitis B virus (HBV) is still the most common etiological factor of HCC.6,7 HBV is a hepatotropic virus that chronically infects approximately 400 million people worldwide and is thought to be responsible for a million deaths annually.8,9 Especially in the People’s Republic of China, HBV infection is a challenging health issue with approximately 93 million people being HBV carriers and 30 million people being chronic hepatitis B patients.10,11 HBV infection is associated with a large spectrum of liver diseases including chronic hepatitis, cirrhosis, and HCC. The pathogenesis of HCC in chronic HBV infections has been investigated comprehensively in recent decades.12–15 Studies12–15 have demonstrated a significant relationship between serum HBV DNA levels and the risk of developing HCC in chronic HBV carriers. In addition to viral load, HBV genotype and mutations are also correlated with the development of HCC.14–17 However, only a few studies have focused on the prognostic value of these mutations in HBV-related HCC (HBV-HCC) patients.18,19
The HBV genome contains four overlapping open reading frames that encode the surface protein (S), the core protein, a polymerase, and a multifunctional nonstructural protein called X.13,20 HBV has a mutation rate that is more than 10-fold higher than that of other DNA viruses due to the deficiency of the proofreading function for HBV polymerase. The HBV mutations are clustered in a particular region of the HBV genome such as the basal core promoter (BCP)/preCore region and the Pre S/S region. The most frequent BCP mutation is a double mutation involving an A to T substitution at nucleotide 1762 and a G to A substitution at nucleotide 1764. The A1762T/G1764A BCP double mutation was associated with a hazard ratio (HR) of 1.73 for developing HCC.12–15 Other BCP mutations, notably T1753V and T1768A, have also been reported to be associated with an increased risk of HCC. The X gene mutations (two of the most common being C1653T and T1753V) and Pre S2 gene deletions have been associated with increased incidence of HCC.13–15 Accumulation of mutations reflects both the activity of virus infection and the strength of the immune response. However, relatively little is known about the association between the genomic changes in HBV DNA and the clinical outcomes of patients with HBV-HCC after surgical resection.
In present study, we sought to determine whether the genomic mutations in HBV DNA are associated with the clinical characteristics and outcomes of HBV-HCC patients after surgical resection.
Materials and methods
Patients and clinical treatment
A cohort of 131 consecutive patients who received hepatectomy for HCC in our department from January 2010 to December 2010 were retrospectively enrolled. The inclusion criteria for the present study were: 1) no previous treatment for HCC before surgery; 2) histologic confirmation of HCC; 3) HBV positive; 4) R0 resection; 5) no lymph node or extrahepatic metastasis; and 6) follow-up period ≥6.0 months. Patients co-infected with immunodeficiency virus (HIV), hepatitis C virus, or hepatitis D virus were excluded. All the preoperative parameters were recorded and evaluated as possible predictors of survival including sex, age, HBV DNA level, platelet count, AFP, total bilirubin level (TBil), albumin, liver function status, tumor size and number, and tumor thrombus. Liver functional reserve was also assessed using Child-Pugh classification.
Hepatectomy was carried out under general anesthesia using a right subcostal incision with a midline extension. Intraoperative ultrasonography was routinely performed to confirm resectability and major vascular structures. Pringle’s maneuver was routinely used with a clamp/unclamp time of 10 minutes/5 minutes. The liver parenchyma was divided with clamp-crushing technique or ultrasonic dissector according to the surgeon’s preference. Anatomic resection was our preferred surgical method in hepatic resection for multiple nodules in one segment or in neighboring segments. For anatomic resection, the hepatic parenchyma was transected at the intersegmental plane as described by Couinaud et al.30 For multiple bilobar nodules, anatomic resection was preferred for the main tumor, while satellite nodules were resected nonanatomically with intent for a negative resection margin. When an inadequate liver remnant was suspected, nonanatomic resection was performed to try to achieve a negative resection margin. Hemostasis on the raw liver surface was done with suturing and fibrin glue.
After surgery, patients were followed up every 3 months for the first 2 years and every 6 months thereafter with physical examination, blood tests for AFP and liver function, and contrast abdominal computed tomography. Anti-virus treatment with lamivudine, entecavir or adefovir was given according to the APASL guideline.6 Causes of death and sites of recurrence were determined from death certificates, medical interviews, and radiological findings. OS was defined as the interval between surgery and time of either death or last follow-up. Disease-free survival (DFS) was defined as the length of time after liver resection for HCC to detectable intrahepatic and/or extrahepatic recurrence. The treatment for recurrent tumors was determined by our multidisciplinary team including surgeons, oncologists, radiologists, hepatologists, and pathologists. The last follow-up date for patients still alive was in December 2016.
Ethics statement
The research was approved by the institutional review board of Sun Yat-sen University Cancer Center, and written informed consent was obtained from each patient involved in the study.
All the procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments, or comparable ethical standards.
Virological assay
The methods have been described previously by others.18,19,21–23 All the samples were collected within 30 minutes after resection; the matched paired tumor and corresponding surrounding non-tumorous liver tissue samples were immediately frozen at −80°C. The Wizard Genomic DNA extraction kit (Promega, Madison, WI, USA) was used to extract genomic DNA in surrounding non-tumorous liver tissue. The ABI 7300 TaqMan platform (Life Technologies, Carlsbad, CA, USA) was used to determine the HBV DNA concentration, and the concentration was calculated as copies per microgram of genomic DNA. The DNA sequences were identified by using fluorescence-labeled primers with 3700 Automatic Sequencer (ABI, Foster City, CA, USA). According to the sequencing outcome, HBV genotype and mutations (including A1752T/G, T1753C, G1757A, A1762T/G1764A, C1766T, T1768A, A1775G, C1799G, A1846T, T1858C, G1896A, G1898A, G1899A, and Pre S deletion) were confirmed by the BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analyses
The statistical analyses were performed using the SPSS 13.0 statistical software (SPSS Company, Chicago, IL, USA). We compared the presence of genomic changes in HBV DNA as well as the clinical characteristics with the clinical outcomes of these patients after surgical resection. Comparisons between the two groups were done using the Student’s t-test for continuous data and the chi square test for categorical data. The OS and DFS were calculated by Kaplan-Meier method and compared by log-rank test. The prognostic variables in predicting OS and DFS were assessed by multivariate Cox proportional hazards regression analysis. Results were given as mean ± SD. All statistical tests were two sided, and a significant difference was considered when p<0.05.
Results
Baseline characteristics
The baseline characteristics of all patients are described in detail in Table 1. In brief, there were 118 male and 13 female patients, with a median age of 50 years (mean ± SD: 49.6±10.5, range: 16–73 years). Tumor size ranged from 1.0 cm to 18.0 cm (mean ± SD: 5.6±3.8); 78 patients (59.5%) had tumor ≤5.0 cm and 53 (40.5%) had tumor >5.0 cm. A total of 99 patients (75.6%) had a single tumor and 32 (24.2%) had multiple tumors. All patients were HBV positive and HIV/HCV/HDV negative. The median HBV DNA was 1.3×105 IU/L, ranging from 0 to 2.57×108 IU/L. According to the 7th edition of the tumor-node-metastasis (TNM) classification of the American Joint Committee on Cancer, 94 patients (71.8%) had stage I–II disease, and 37 (28.2%) had stage III disease (Table 1).
Anatomic resections based on the segments defined by Couinaud et al30 were performed in 72.5% of patients (95/131). Major hepatectomy, defined as resection of more than two anatomic segments, was required in 68.7% of patients (80/131). The mean operative time was 126.7±37.5 minutes (interquartile range [IQR] 90–240); the mean blood loss was 147.5±35.5 mL (IQR 50–1500). The median length of hospital stay after surgery was 7 days (IQR 6–15). In all, 36 of 131 (27.5%) patients developed postoperative complications. The 30-day mortality rate was 0%.
Prevalence of HBV genotype and genomic mutations
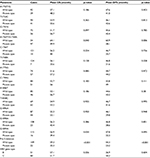
Among the 131 cases, 51 cases (38.9%) were HBV genotype type B and 80 (61.1%) were type C. Most mutation regions that have been reported (including Pre S1, Pre S2, part of S region, part of X region, BCP region, part of C region) were covered in the sequencing. According to different mutation regions, A1752T/G, T1753C, G1757A, A1762T/G1764A, C1766T, T1768A, A1775G, C1799G, A1846T, T1858C, G1896A, G1898A, G1899A, and Pre S deletion mutations were recognized in 9.8% (39/131), 31.3% (41/131), 42.7% (56/131), 74.0% (97/131), 7.6% (10/131), 5.3% (7/131), 43.5% (57/131), 38.9% (51/131), 35.1% (46/131), 47.3% (62/131), 33.6% (44/131), 17.6% (23/131), 12.2% (16/131), and 16.8% (22/131) cases, respectively (Table 2).
  | Table 2 HBV genomic mutations and the relation to patient survival Abbreviations: DFS, disease-free survival; HBV, hepatitis B virus; OS, overall survival. |
HBV genomic mutations related to survival and clinical characteristics
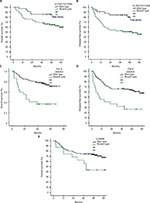
As shown in Table 2, most of the HBV genomic mutations (including A1752T/G, T1753C, G1757A, C1766T, T1768A, A1775G, C1799G, A1846T, T1858C, G1896A, and G1898A) did not make a significant statistical difference in OS and DFS. However, A1762T/G1764A mutation related to worse OS and DFS (p=0.040 and p=0.006, respectively), G1899A mutation related to worse OS (p=0.030), and Pre S deletion mutation related to worse OS and DFS (p<0.001 and p<0.001, respectively, Table 2, Figure 1).
The relationships between A1762T/G1764A, G1899A, and Pre S deletion mutations and clinical parameters were analyzed respectively. As shown in Tables S1, S2, and S3, the A1762T/G1764A mutation is correlated with tumor size (r=0.204, p=0.019), G1899A mutation with vascular invasion (r=0.332, p<0.001), and Pre S deletion with AFP (r=0.254, p=0.003), positively.
A1762T/G1764A mutation and Pre S deletion predict survival
The median follow-up period was 55.0 months. The 1-, 3-, and 5-year OS was 84%, 73.3%, and 64.9%, respectively. The 1-, 3-, and 5-year DFS was 71.8%, 57.3%, and 48.3% respectively.
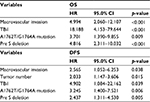
The univariate and multivariate analyses of prognostic factors for OS and DFS were analyzed (Tables 1–3). All clinical parameters were included in univariate analysis for OS and DFS (Table 1). All variables with p<0.10 in univariate analysis, as well as A1762T/G1764A, G1899A and Pre S deletion mutations, were further included in multivariate analysis with Cox proportional hazards model. As shown in Table 3, macrovascular invasion (HR=4.994, 95% CI 2.060–12.107, p<0.001), TBil level (HR=18.188, 95% CI 4.153–79.644, p<0.001), A1762T/G1764A mutation (HR=3.701, 95% CI 1.390–9.855, p=0.009), and Pre S deletion (HR=4.816, 95% CI 2.311–10.032, p<0.001) were independent prognostic factors for OS. Macrovascular invasion (HR=2.656, 95% CI 1.052–6.253, p=0.038), tumor number (HR=2.033, 95% CI 1.147–3.606, p=0.015), TBil level (HR=4.902, 95% CI 1.084–22.162, p=0.039), A1762T/G1764A mutation (HR=3.245, 95% CI 1.400–7.521, p=0.006), and Pre S deletion (HR=2.437 95% CI 1.311–4.530, p<0.001) were independent prognostic factors for DFS.
  | Table 3 Multivariate analyses of prognostic factors for OS and DFS Abbreviations: DFS, disease-free survival; HR, hazard ratio; OS, overall survival; TBil, total bilirubin level. |
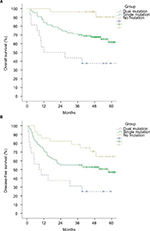
Since both A1762T/G1764A mutation and Pre S deletion were independent prognostic factors for OS and DFS, the 131 patients were divided into three groups according to A1762T/G1764A mutation and Pre S deletion status: no mutation (26 cases), single mutation (77 cases), and dual mutations (16 cases). The baseline clinical characteristics are listed in Table S4, and there were no significant differences between groups. As it was shown in Figure 2, the dual mutation group related to the worst OS and DFS, then the single mutation, and lastly the no mutation group (p<0.001 and p<0.001, respectively).
A1762T/G1764A mutation and Pre S deletion related to early recurrence
During the follow-up, a total of 74 patients experienced tumor recurrence, 54 cases within 2 years (early recurrence) and 20 cases after 2 years (late recurrence). As shown in Table 4, patients with A1762T/G1764A mutation or Pre S deletion were more likely to have early recurrence (p=0.042 and p=0.019, respectively). Especially for patients with A1762T/G1764A and Pre S deletion dual mutation, the early recurrence rate (62.5%, 10/16) was much higher than for those with single mutation (44.8%, 39/87) or no mutation (17.9%, 5/28, p=0.008). No relationships can be identified between A1762T/G1764A mutation and/or Pre S deletion with late recurrence (data not shown).
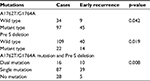  | Table 4 A1762T/G1764A mutation and Pre S deletion relate to early recurrence |
Discussion
Although HBV is still one of the most common etiological factors of HCC, relatively little is known about the association between the HBV genomic mutations and the clinical characteristics and outcomes of HBV-HCC patients after surgical resection.22,24 In the present study, most mutation regions that have been reported in the literature previously, were sequenced,20,23 and the results demonstrated that HBV DNA genomic mutations in A1762T/G1764A and Pre S deletion were associated with worse prognosis and early recurrence for HBV-HCC patients after surgery. Our results highlight the importance of anti-virus treatment for HBV-HCC after surgery.
A few studies have focused on the prognostic value of the HBV genomic mutations in HBV-HCC patients, but the results remain controversial. By assessing HBV DNA in liver tissue specimens, in 2010, Yeh et al25 from Taiwan showed for the first time that the BCP A1762T/G1764A mutation is independently predictive of postoperative survival in HCC patients. In 2011, Ryu et al26 from Korea reported that Pre S deletion and BCP mutations in HBV of genotype C2 did not affect the initial tumor features, but Pre S deletion was an independent risk factor affecting HCC survival. However, in 2013 Mathews et al23 from Korea revealed that the postoperative recurrence or survival was not affected by the genomic changes at the preCore, BCP, X, and Pre S2 regions in HBV of genotype C2 in patients with HBV-HCC treated with curative surgical resection, by assessing serum HBV DNA in a total of 247 patients. Su et al19 from Taiwan demonstrated that among patients with higher serum HBV DNA levels, those with Pre S deletion had significantly higher rates of recurrence after surgery. More recently, Guo et al18,21 from People’s Republic of China identified that HBV-HCC survival after surgery is associated with HBV mutations in the X protein region, preCore regions, and small S genes. In the present study, we sequenced most mutation regions that have been reported in the literature previously, and found that both A1762T/G1764A and Pre S deletion mutations related to worse OS and DFS. Compared to previous studies, our study has the following advantages: 1) our study covered the 14 mutation regions, more regions than previous studies; 2) both genotype B and C HBV-HCC, which are common in East Asia, were included in our study; 3) the median follow-up period was 55.0 months, which is sufficient; and 4) we detected the HBV DNA directly from liver tissue, which is believed to be more reliable.
The A1762T/G1764A double mutation is the most frequent BCP mutation. It downregulates HBeAg expression, promotes immunological tolerance, accelerates HBV copying and HBsAg expression, and finally leads to serious liver damage.20,27 Previous studies23,25 had confirmed that A1762T/G1764A double mutation is not only an independent risk factor for HCC but also a predictor for poor survival in HBV-HCC patients. Patients with A1762T/G1764A double mutation tend to have poor liver function and serious cirrhosis, which will reduce time of survival after surgery. In the present study, we also revealed that the A1762T/G1764A mutation is correlated with tumor size, which indicates that A1762T/G1764A mutation might accelerate tumor proliferation through some unknown mechanism. More recently, Chen et al22 reported that A1762T/G1764A double mutation and phosphorylated AKT (pAKT) correlated with proliferation and microvascularization but inversely correlated with apoptosis in HCC samples; levels of p21 and p27 were decreased in A1762T/G1764A double mutation or pAKT overexpressing HCC due to SKP2 upregulation; levels of E2F1, and both mRNA and protein of SKP2, were increased in A1762T/G1764A double mutation HCC. Further studies are needed to uncover the underlying mechanism.
The Pre S region of HBV is crucial in mediating the attachment of the virus to host hepatocytes.20 Moreover, it is important for interacting with the host immune responses. It has been reported that the proportions of Pre S deletion mutants are significantly higher in patients with HBV-related HCC than in those with inactive HBV carrier status, implying that these mutants may be associated with HCC carcinogenesis.15 Su et al19 reported that patients with Pre S deletion mutants have a higher proportion of positive HBeAg in sera and a trend of higher HBV DNA levels than their counterparts; the presence of Pre S deletion mutants correlates to a higher rate of tumor recurrence in patients with serum HBV DNA levels >105 copies/mL or HBsAg >500 IU/mL. However, the correlations cannot be observed in patients with low viral loads or in those receiving antiviral therapy after resection. It implies that the hepatocarcinogenesis and tumor recurrence induced by Pre S deletion mutants may be based on active HBV viral replication, the accumulation of oxidative stress, and subsequent chromosomal instability.19
More importantly, for the first time, we demonstrated that HBV-HCC with dual mutations in A1762T/G1764A and Pre S deletion related to the worst OS and DFS, and patients tended to have early recurrence. Accumulation of mutations reflects both the activity of virus infection and the strength of the immune response.22 Previous studies15 have demonstrated that the accumulated mutations in HBV DNA increase the risk to develop HCC in chronic HBV carriers. Huang et al28 reported that the combo mutant accelerated p21 (WAF1/CIP1) degradation by the upregulated expression of SKP2, and increased cyclin E expression in primary hepatocytes and HepG2 cells, resulting in the combo mutant accelerating cell cycle progression. Chen et al22 also reported that the accumulation of A1762T/G1764A combined with other mutation(s) was more common in patients with poor prognosis than in patients with better prognosis. But the underlying mechanism remains unclear. However, these findings highlight the importance of anti-HBV treatment for the HBV-HCC patients. HBV infection contributes to not only hepatocarcinogenesis and tumor progression but also tumor recurrence. Clinical trials29 have shown that patients who receive anti-virus treatment have significantly decreased early recurrence and improved liver function 6 months after surgery, and anti-virus treatment significantly improved both OS and DFS. Our findings also indicated that more aggressive anti-HBV therapy and more frequent follow-up should be suggested for those HBV-HCC patients with dual mutations in A1762T/G1764A and Pre S deletion. However, our findings should be verified by prospective studies.
Other HBV mutations, which have been reported to relate to liver cirrhosis and HCC, including A1752T/G, T1753C, G1757A, C1766T, T1768A, A1775G, C1799G, A1846T, T1858C, and G1898A, failed to show the association with patients’ prognosis in the present study. Therefore, no further analysis was performed.
There are several limitations in the present study that must be taken into account. Firstly, sample size was relatively small with only 131 patients included, although the follow-up period was relatively long. Secondly, HBV DNA in serum was unavailable in the present study and the relationship between liver tissue HBV DNA and serum HBV DNA cannot be analyzed. Thirdly, the HBV DNA statuses in recurrent tumors are not available.
Conclusion
The present study demonstrated that HBV DNA genomic mutations in A1762T/G1764A and Pre S deletion were associated with worse prognosis and early recurrence for HBV-HCC patients after surgery. Anti-virus treatment might benefit HBV-HCC patients.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (NSFC81572387 and 81602143), and the Scientific Project of Guangdong Province (2017A020215034).
Disclosure
The authors report no conflicts of interest in this work.
References
Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. | ||
Khalaf N, Ying J, Mittal S, et al. Natural history of untreated hepatocellular carcinoma in a US cohort and the role of cancer surveillance. Clin Gastroenterol Hepatol. 2017;15(2):273–281.e1. | ||
Dhir M, Melin AA, Douaiher J, et al. A review and update of treatment options and controversies in the management of hepatocellular carcinoma. Ann Surg. 2016;263(6):1112–1125. | ||
Poon RT, Cheung TT, Kwok PC, et al. Hong Kong consensus recommendations on the management of hepatocellular carcinoma. Liver Cancer. 2015;4(1):51–69. | ||
de Lope CR, Tremosini S, Forner A, Reig M, Bruix J. Management of HCC. J Hepatol. 2012;56 Suppl 1:S75–S87. | ||
Sarin SK, Kumar M, Lau GK, et al. Asian-Pacific clinical practice guidelines on the management of hepatitis B: a 2015 update. Hepatol Int. 2016;10(1):1–98. | ||
El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–1273.e1. | ||
Sherman M, Bruix J, Porayko M, Tran T; AASLD Practice Guidelines Committee. Screening for hepatocellular carcinoma: the rationale for the American Association for the Study of Liver Diseases recommendations. Hepatology. 2012;56(3):793–796. | ||
Chiang CJ, Yang YW, You SL, Lai MS, Chen CJ. Thirty-year outcomes of the national hepatitis B immunization program in Taiwan. JAMA. 2013;310(9):974–976. | ||
Di Bisceglie AM. Hepatitis B and hepatocellular carcinoma. Hepatology. 2009;49(5 Suppl):S56–S60. | ||
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. | ||
Liu S, Zhang H, Gu C, et al. Associations between hepatitis B virus mutations and the risk of hepatocellular carcinoma: a meta-analysis. J Natl Cancer Inst. 2009;101(15):1066–1082. | ||
Zhang Q, Cao G. Genotypes, mutations, and viral load of hepatitis B virus and the risk of hepatocellular carcinoma. HBV properties and hepatocarcinogenesis. Hepat Mon. 2011;11(2):86–91. | ||
Yang HI, Yeh SH, Chen PJ, et al; REVEAL-HBV Study Group. Associations between hepatitis B virus genotype and mutants and the risk of hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(16):1134–1143. | ||
Llovet JM, Lok A. Hepatitis B virus genotype and mutants: risk factors for hepatocellular carcinoma. J Natl Cancer Inst. 2008;100(16):1121–1123. | ||
Yeh CT, Chen T, Hsu CW, et al. Emergence of the rtA181T/sW172* mutant increased the risk of hepatoma occurrence in patients with lamivudine-resistant chronic hepatitis B. BMC Cancer. 2011;11:398. | ||
Chan HL, Tse CH, Mo F, et al. High viral load and hepatitis B virus subgenotype ce are associated with increased risk of hepatocellular carcinoma. J Clin Oncol. 2008;26(2):177–182. | ||
Xie Y, Liu S, Zhao Y, et al. Precore/core region mutations in hepatitis B virus DNA predict postoperative survival in hepatocellular carcinoma. PLoS One. 2015;10(7):e0133393. | ||
Su CW, Chiou YW, Tsai YH, et al. The influence of hepatitis B viral load and Pre-S deletion mutations on post-operative recurrence of hepatocellular carcinoma and the tertiary preventive effects by anti-viral therapy. PLoS One. 2013;8(6):e66457. | ||
Zhang ZH, Wu CC, Chen XW, Li X, Li J, Lu MJ. Genetic variation of hepatitis B virus and its significance for pathogenesis. World J Gastroenterol. 2016;22(1):126–144. | ||
Peng L, Yang G, Wu C, Wang W, Wu J, Guo Z. Mutations in hepatitis B virus small s genes predict postoperative survival in hepatocellular carcinoma. Onco Targets Ther. 2016;9:7367–7372. eCollection 2016. | ||
Chen L, Gu L, Gu Y, et al. HBV core promoter mutations and AKT upregulate S-phase kinase-associated protein 2 to promote postoperative hepatocellular carcinoma progression. Sci Rep. 2016;6:35917. | ||
Mathews P, Lee D, Chung Y, et al. Effects of genomic changes in hepatitis B virus on postoperative recurrence and survival in patients with hepatocellular carcinoma. Ann Surg Oncol. 2013;20(4):1216–1222. | ||
Jang JW, Chun JY, Park YM, et al. Mutational complex genotype of the hepatitis B virus X /precore regions as a novel predictive marker for hepatocellular carcinoma. Cancer Sci. 2012;103(2):296–304. | ||
Yeh CT, So M, Ng J, et al. Hepatitis B virus-DNA level and basal core promoter A1762T/G1764A mutation in liver tissue independently predict postoperative survival in hepatocellular carcinoma. Hepatology. 2010;52(6):1922–1933. | ||
Ryu HJ, Kim DY, Park JY, et al. Clinical features and prognosis of hepatocellular carcinoma with respect to pre-S deletion and basal core promoter mutations of hepatitis B virus Genotype C2. J Med Virol. 2011;83(12):2088–2095. | ||
Tseng TC, Liu CJ, Yang HC, et al. High levels of hepatitis B surface antigen increase risk of hepatocellular carcinoma in patients with low HBV load. Gastroenterology. 2012;142(5):1140–1149.e3; quiz e13–e14. | ||
Huang Y, Tong S, Tai AW, Hussain M, Lok AS. Hepatitis B virus core promoter mutations contribute to hepatocarcinogenesis by deregulating SKP2 and its target, p21. Gastroenterology. 2011;141(4):1412–1421, 1421.e1–e5. | ||
Yin J, Li N, Han Y, et al. Effect of antiviral treatment with nucleotide/nucleoside analogs on postoperative prognosis of hepatitis B virus-related hepatocellular carcinoma: a two-stage longitudinal clinical study. J Clin Oncol. 2013;31(29):3647–3655. | ||
Couinaud C, Delmas A, Patel J. Le foie: études anatomiques et chirugicales. [The liver: anatomical and surgical studies]. Paris: Masson & Cie; 1957. French |
Supplementary materials
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.