Back to Journals » OncoTargets and Therapy » Volume 13
The microRNA miR-3174 Suppresses the Expression of ADAM15 and Inhibits the Proliferation of Patient-Derived Bladder Cancer Cells
Authors Yu C, Wang Y, Liu T, Sha K, Song Z, Zhao M, Wang X
Received 20 January 2020
Accepted for publication 18 April 2020
Published 14 May 2020 Volume 2020:13 Pages 4157—4168
DOI https://doi.org/10.2147/OTT.S246710
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Chunhu Yu,1 Ying Wang,1 Tiejun Liu,1 Kefu Sha,1 Zhaoxia Song,1 Mingjun Zhao,1 Xiaolin Wang2
1Department of Urinary Surgery, Beijing Rehabilitation Hospital of Capital Medical University, Beijing 100144, People’s Republic of China; 2The Third District of Airforce Special Service Sanatorium, Chinese People’s Liberation Army Air Force, Hangzhou 310021, Zhejiang Province, People’s Republic of China
Correspondence: Xiaolin Wang Email [email protected]
Background: Bladder cancer is a major urinary system cancer, and its mechanism of action regarding its progression is unclear. The goal of this study was to examine the expression of ADAM panel in the clinical specimens of bladder cancer and to investigate the role of miR-3174/ADAM15 (a disintegrin and metalloprotease 15) axis in the regulation of bladder cancer cell proliferation.
Methods: The expression of an ADAM gene panel (including ADAM8, 9, 10, 11, 12, 15, 17, 19, 22, 23, 28, and 33), including 30 pairs of bladder tumor and non-tumor specimens, was examined by Ion AmpliSeq Targeted Sequencing. A microRNA (miRNA) that could potentially target the ADAM with the highest expression level in the tumor tissue was identified using the online tool miRDB. Next, the interaction between the miRNA and ADAM15 was identified by Western blot. Finally, the proliferation of bladder cancer cells was examined using MTT (3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide) experiments (cell proliferation examining) and subcutaneous tumor models by using nude mice.
Results: The expression of ADAM15 in tumor tissue was found statistically significant when compared to its expression in non-tumor tissue. Additionally, ADAM15’s expression in tumor tissue was found the highest of all other tested ADAMs. Next, by using the online tool miRDB, a microRNA termed miR-3174 was identified that targets ADAM15 and inhibits its expression by binding to its 3′-untranslated region. Finally, we found that overexpression of miR-3174 in bladder cancer cells inhibited the proliferation of cells due to the inhibition of ADAM15.
Conclusion: In the present work, the data highlight that miR-3174 inhibits the proliferation of bladder cancer cells by targeting ADAM15.
Keywords: ADAM, bladder cancer, metastasis, microRNA, proliferation
Introduction
Bladder cancer is one of the major urinary system cancers; however, the details of its mechanisms, such as cell proliferation and metastasis, remain scarce.1–3 ADAMs (a disintegrin and metalloproteases), which belong to a protein family of de-integrating metalloproteinases, are type I transmembrane proteins (zinc-containing proteases) and play important roles in regulating cell-cell and cell-matrix interactions.4 So far, more than 30 types of ADAMs have been identified.4,5 The structure of an ADAM is featured by a conserved RGD (Arg-Gly-Asp) motif in its integrin domain that regulates cell-cell adhesion.5,6 ADAMs have also been found to regulate various physical processes of the cell, including proteolysis, cell fusion, migration, signal transduction, and release of biological activity factors.6 Recently, ADAMs have started to be considered as important regulators of human cancers, including lung, gastric, colon, breast, and pancreatic cancer.7 In the present study, we demonstrate that among 12 types of ADAMs, the expression of ADAM15 was found the highest in bladder cancer tissue. Here, miR-3174 was found as a microRNA that targets ADAM15. Moreover, the expression of miR-3174 was found to inhibit the expression of ADAM15 in bladder cancer tissue. As a result, overexpression of miR-3174 inhibited the cell proliferation, metastasis, and subcutaneous growth in vivo of patient-derived bladder cancer cells.
Materials and Methods
Patient-Derived Bladder Cancer Cell Lines
The paired tissues (the bladder cancer tissues or the non-tumor tissues obtained in the same patient) of bladder tumor and non-tumor specimens were obtained by surgery. The collection of clinical specimens was in compliance with the Declaration of Helsinki, and the pathological stages (Class 0, A, B, C and D) of bladder cancer tissues were measured using the Jewett-Marshall methods.8–10 Furthermore, this study and its applied methods were approved by the ethics committee of the Beijing Rehabilitation Hospital, which is affiliated to the Capital Medical University (Beijing, China). Written informed consent was obtained from all the patients. The surgically resected bladder cancer specimens were preserved in Dulbecco’s Modified Eagle Medium (DMEM, Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 20% fetal bovine serum (FBS, Thermo Fisher Scientific). Then, to obtain a patient-derived tumor cell line, the tissue was ground with a 200-mesh steel sieve, and the cell suspension washed with PBS. Next, the single cells derived from clinical specimens of bladder cancer tissues were cultured in DMEM with 10% FBS. There were five lines of PDCs including in the present work, and the PDCs included two lines of Class C (No. 1 and No. 2) and three lines of Class D (No. 3, 4 and 5).
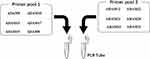
Ion AmpliSeq Targeted Sequencing
Amplification primers for the ADAM8, ADAM9, ADAM10, ADAM11, ADAM12, ADAM15, ADAM17, ADAM19, ADAM22, ADAM23, ADAM28, and ADAM33 genes were designed using the Ion AmpliSeq Designer tool from Thermo Fisher Scientific.11 Before use, the primers were mixed together into one primer pool. For testing samples, the total RNA was extracted from each bladder cancer cell line using an RNA-extraction Kit (Applied Biosystems instruments, from Thermo Fisher Scientific). These samples were then subjected to reverse transcription by using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Next, the library construction of cDNA was performed using the Ion AmpliSeq Library Kit and the Ion Xpress Barcode Adapter reagent (Life Technologies, USA). The DNA targets—including the ADAM gene panel—were amplified using a 2 × primer pool buffer with 22 cycles’ amplify (Ion PGM Hi-Q View Sequencing Kit, Catalog number: A30044, Life Technologiesy Corporation, USA). The Ion PGM Hi-Q sequencing kit and the Ion Torrent 318 chip (Ion 318 Chip Kit v2 BC, Catalog number: 4488150, Life Technologiesy Corporation, USA) were used for sequencing on the Ion Torrent PGM platform (Ion Personal Genome Machine [PGM] System, Catalog number: 4462921, Life Technologiesy Corporation, USA). Finally, the Ion Torrent data were analyzed using the Ion Torrent Suite v3.0 software (Supplemental Table 1).
Quantitative Polymerase Chain Reaction (qPCR)
Total RNA samples were extracted from bladder cancer cells by using an RNA-extraction Kit (Applied Biosystems Instruments, from Thermo Fisher Scientific), and the RNA samples were reverse transcribed into cDNA by using an RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. The kit (miRNA examination kit) to examine the expression level of miR-3174 was purchased from Thermo Fisher Scientific. Next, the qPCR experiments were performed following the manufacturer’s instructions (Applied Biosystems Instruments, from Thermo Fisher Scientific) and the primers from Table 1. The level of GAPDH mRNA was used as an internal control. The expression of ADAM15 and miR-3174 in bladder cancer specimens was processed into a scatter plot image: The x-axis represented the mRNA level of ADAM15 and the y-axis the miR-3174 level, both from the same sample.
 |
Table 1 The Inhibitory Rates of miR-3174 on Five Patient-Derived Bladder Cancer Cells’ Proliferation |
Plasmids and Lentivirus Particles
The expression vectors containing full-length sequences of pri-muR-3174 and ADAM15 were purchased from Vigene Biosciences (Jinan City, Shandong Province, China). The lentivirus particles containing full-length sequences of pri-muR-3174, wild type ADAM15, and ADAM15 with mutated miR-3174 binding sites were also prepared by Vigene.
Cell Culture and Proliferation
The patient-derived bladder cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific). The cells were treated with lentivirus particles to introduce the expression of miR-3174 or ADAM15. Then, for the proliferation experiments, 2000 cells were seeded per well into a 96-well cell culture plate. After 3 days of culture, the cells were analyzed by MTT analysis following the manufacturer’s instructions (Ameresco, USA). The inhibition rates were calculated as [(O.D. [optical density] 490 nm of the control group at day 3 of culture – O.D. 490 nm of the control group at day 0 of culture) – (O.D. 490 nm of the administration group at day 3 of culture – O.D. 490 nm of the administration group at day 0 of culture)]/(O.D. 490 nm of the control group at day 3 of culture – O.D. 490 nm of the control group at day 0 of culture) × 100%.12,13
Transwell Experiments
The patient-derived bladder cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific). The cells were treated with lentivirus particles to introduce the expression of miR-3174 or ADAM15. For the cell invasion transwell experiment, the bottom of the chamber was first coated with ECM gel (extra-cellular matrix gel, Sigma-Aldrich, St. Louis, USA). For the cell migration transwell experiment, the ECM gel was not required. The cells were seeded into the transwell chambers (8000 cells per chamber). For both experiments, the chambers were cultured for 12–16 hours at 37°C with 5% CO2 condition. Next, the chambers were fixed using absolute ethanol, followed by staining with a crystal violet dye solution (0.25% w/v). Afterward, images of the chambers were obtained, and the chambers extracted by absolute ethanol. The absorbance value (O.D., optical density) was measured at a wavelength of 546 nm. The inhibition rates were calculated as (O.D. 546 nm of the control group – O.D. 546 nm of the administration group)/(O.D. 546 nm of the control group) × 100%.14–16
Subcutaneous Tumor Model
The patient-derived bladder cells were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Thermo Fisher Scientific). The cells were treated with lentivirus particles to introduce the expression of miR-3174 or ADAM15. The subcutaneous in vivo tumor model experiments were reviewed and approved by the Institutional Animal Care and Use Committee of the 302nd Hospital of People’s Liberation Army (Beijing, People’s Republic of China). Nude mice (4–6 weeks old) were purchased from Si-Bei-Fu Biotechnology (Beijing, People’s Republic of China). All animal studies were performed in accordance with the UK (United Kingdom) Animals (Scientific Procedures) Act 1986 and associated guidelines. The cells were injected into a subcutaneous position in the mice. Next, the mice were sacrificed after 6 to 8 weeks, and the subcutaneous tumors were collected (n = 10 for per group). The mass of the tumors was measured using a precise balance. The length and width of the subcutaneous tumors were measured by a vernier caliper, and the volumes of subcutaneous tumors were calculated as (tumor length × tumor width × tumor width)/2.17–19 The inhibition rates were calculated as (tumor volume of the control group tissue – tumor volume of the administrated group tissue)/(tumor volume of the control group tissue) × 100% and as (tumor weight of the control group tissue – tumor weight of the administrated group tissue)/(tumor weight of the control group tissues) × 100%.20
Statistical Analysis
Analyses to determine the statistical significance of the obtained data were performed using GraphPad Software (version 6.0, San Diego, CA. USA). The statistical significance between the two groups was analyzed using the two-way ANOVA (Analysis of Variance) with a Bonferroni correction. Additionally, the correlation analysis of the expression of miR-3174 or ADAm15 was performed using linear regression methods. Paired samples were tested using the paired-sample t-test. All values with a P-value <0.05 were concerned statistically significant.
Results
The Expression of ADAM15 in Bladder Tumor Tissue Was Significantly Higher Compared to the Paired Non-Tumor Tissue
First, the expression level of all ADAMs (ADAM8, 9, 10, 11, 12, 15, 17, 19, 22, 23, 28, and 33) was examined in 30 pairs of clinical specimens (one cancer and one non-cancer as one pair) from bladder cancer patients. The results show that the expression of ADAM23 in tumor tissue was lower than that in non-tumor tissue (Figure 1A). The expression of ADAM11 (Figure 1K), ADAM28 (Figure 1B), and ADAM33 (Figure 1C) was slightly lower in tumor tissue compared to non-tumor tissue; nevertheless, no statistical difference was found. For ADAM17 (Figure 1E), ADAM15 (Figure 1F), ADAM12 (Figure 1G), and ADAM10 (Figure 1H), the expression was significantly higher in tumor tissue than in non-tumor tissue (Figure 1). This, while the expression of ADAM8 (Figure 1J), ADAM9 (Figure 1I), ADAM19 (Figure 1L), and ADAM22 (Figure 1D) was basically the same between the tumor and non-tumor tissue.
Compared to the expression level of different ADAMs in the tumor tissue, the expression level of ADAM15 was significantly higher (Figure 2). A correlation analysis was performed of ADAM15’s expression in the tumor as well as non-tumor tissue. As a result, the expression of ADAM15 was found to be inversely correlated (y =0 − 0.0918x + 0.0252; R2 = 0.2862; P = 0.0023) in both the tumor and non-tumor tissue (Figure 3A). To further examine the correlation between the expression of ADAM15 and various stages of bladder cancer, the expression level of ADAM15 was evaluated in various stages: Class 0, A, B, C, and D (Figure 3B). The results show that a high level of ADAM15 is present in more aggressive stages of bladder cancer (eg, Class C and D) than in less aggressive stages of bladder cancer (eg, Class 0 or Class A) (Figure 3B). Overall, ADAM15 was selected for further investigation.
The microRNA miR‐3174 Targets ADAM15
Next, miRDB was used to identify a microRNA (miR) that potentially binds to ADAM15. As shown in Figure 4A, miR-3174 was identified to have a potential binding site in the 3’UTR (3’ un-transcription region) of ADAM15. To confirm, Western blot analysis was used to determine the effect of miR-3174 on ADAM15’s expression in bladder cancer cells (Figure 4B). The results showed that the transduction of lentivirus containing miR-3174 significantly decreased the expression of ADAM15 (Figure 4B). Transfection of miR-3174’s inhibitor or the expression vector of ADAM15 with a mutated miR-3174 targeting site in 3’UTR almost fully blocked the inhibiting effect of miR-3174 on ADAM15’ expression in bladder cancer cells (Figure 4B). Furthermore, in agreement with the inhibitory effect, expression of miR-3174 was negatively associated with the expression of the ADAM15 protein in the bladder cancer samples (y = 0 − 0.0659x + 0.0225, R2 = 0.1474, P = 0.0362, Figure 4C). Therefore, miR‐3174 was found to be a microRNA that targets ADAM15.
miR‐3174 Reduced the Proliferation of Bladder Cancer Cells by Targeting ADAM15
The effect of miR-3174 on the proliferation of bladder cancer cells was determined using MTT experiments. As shown in Table 1, the transduction of lentivirus containing miR-3174 inhibited the proliferation of bladder cancer cells. The transfection of the miR-3174 inhibitor or the expression vector of ADAM15 with a mutated miR-3174 targeting site in 3’UTR almost fully stopped the inhibiting effect of miR-3174. For these results, the mean ± SD is shown of the inhibition rates of miR-3174 on five patient-derived bladder cancer cell lines (Table 1). Together, these results indicate that miR‐3174 reduced the proliferation of bladder cancer cells by targeting ADAM15.
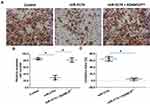
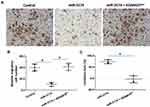
miR‐3174 Inhibits the in vitro Invasion and Migration of Bladder Cancer Cells by Targeting ADAM15
The effect of miR-3174 on the proliferation of bladder cancer cells in vitro was determined using MTT experiments. As shown in Figures 5 and 6, Tables 2 and 3, the transduction of lentivirus containing miR-3174 inhibited the in vitro invasion (Figure 5 and Table 2) and migration (Figure 6 and Table 3) of bladder cancer cells. Similarly, the transfection of the miR-3174 inhibitor or the expression vector of ADAM15 with a mutated miR-3174 targeting site in 3’UTR almost fully stopped the inhibiting effect of miR-3174. The results were found as typical images (Figures 5 and 6) and the mean ± SD of the inhibition rates (Tables 2 and 3) of miR-3174 on five patient-derived bladder cancer cell lines. In sum, miR‐3174 inhibited the in vitro invasion and migration of the bladder cancer cells by targeting ADAM15.
 |
Table 2 The Inhibitory Rates of miR-3174 on Five Patient-Derived Bladder Cancer Cells’ in vitro Invasion |
 |
Table 3 The Inhibitory Rates of miR-3174 on Five Patient-Derived Bladder Cancer Cells’ in vitro Migration |
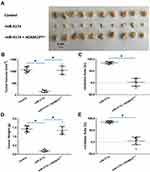
miR‐3174 Inhibits the in vivo Invasion and Migration in Mice of Bladder Cancer Cells by Targeting ADAM15
To further examine the effect of miR-3174 on the subcutaneous growth of bladder cancer cells, nude SCID mice were used as an in vivo model. As shown in Figure 7, Tables 4 and 5, the transduction of lentivirus containing miR-3174 inhibited the subcutaneous growth (Figure 7) of bladder cancer cells. The transduction of lentivirus containing ADAM15, containing a mutated miR-3174 targeting site in 3’UTR, almost fully stopped the inhibiting effect of miR-3174. The results are shown as typical images of subcutaneous tumors (Figure 7) and the mean ± SD of the inhibition rates (Tables 4 and 5) of miR-3174 on five patient-derived bladder cancer cell lines. As such, it can be derived that miR‐3174 reduced the subcutaneous growth of bladder cancer cells in mice by targeting ADAM15.
 |
Table 4 The Inhibitory Rates of miR-3174 on Five Patient-Derived Bladder Cancer Cells’ Subcutaneous Tumor Volumes |
 |
Table 5 The Inhibitory Rates of miR-3174 on Five Patient-Derived Bladder Cancer Cells’ Subcutaneous Tumor Weights |
Discussion
Recently, bladder cancer has been considered as a fatal urinary system disease associated with high metastasis and mortality rates when no optimal treatment is received.21 Although the awareness of hematuria (presence of red blood cells in urine) is still the foremost indicator of bladder cancer, the current early diagnosis of bladder cancer is still not satisfactory.22,23 The prognosis of patients suffering from non-muscle-invasive bladder cancer is relatively good; however, the clinical outcome of bladder cancer with muscle-invasion remains poor.24,25 Thus, it is urgent to research the involvement of metastasis/invasion-related mechanisms in bladder cancer. In the present work, we used Ion AmpliSeq Targeted Sequencing technology to detect the expression levels of ADAMs in 30 pairs of tumor versus non-tumor tissues (Figure 8). Here, the results show that the expression of ADAM15 was significantly higher in tumor tissue than in non-tumor tissue. At the same time, among the 12 tested ADAMs, the expression of ADAM15 was significantly higher in tumor tissue compared to the other ADAMs. The expression of ADAM15 was found to be closely related to the stages of bladder cancer as well. This finding suggests that ADAM15 might play an important role in the occurrence and progression of bladder cancer. The AmpliSeq Targeted Sequencing technology used in the present work is a highly valuable technology. It has several advances: (1) AmpliSeq Targeted Sequencing allows the simultaneous PCR reactions of thousands and even tens of thousands of reactions; (2) the minimum amount of required starting DNA is only 10 ng, which is suitable for the trace specimens; and (3) it is suitable for samples with degraded DNA or RNA due to the long-term cryopreservation or multiple freeze-thaw. So, in our study, the AmpliSeq Targeted Sequencing technology was used to detect the expression of ADAMs in a series of bladder cancer tissue specimens. As a result, the findings of these analyses lay a solid foundation for further study of the role and regulation mechanism of ADAMs in bladder cancer.
Previous studies have confirmed that ADAM is a novel regulator of malignant tumor metastasis and invasion;26 however, similar reports regarding its role in bladder cancer are few and the effects not completely clear.24 In the present work, the expression of various ADAMs in bladder cancer tissue was examined for the first time. From these results, miR-3174 was identified as a miRNA that targets the 3’UTR of ADAM15. Interestingly, overexpression of miR-3174 inhibited the proliferation and metastasis of bladder cancer cells by targeting and inhibiting ADAM15. Based on the factor that expression of miRNAs in human cancer cells could be used to silence oncogene expression, it is a promising approach to achieve an antitumor effect.27–29 Thus, our data reveal not only the role of the ADAM15/miR-3174 axis in bladder cancer regulation but also provides evidence that miR-3174 could be a potential approach for bladder cancer treatment. Moreover, previous work often focused on the genetic alteration of tumorigenesis-associated genes (eg, TP53, VEGFR, and EGFR).1–5 For this reason, our results are highly valuable as it expands the understanding of the molecular mechanisms involved with bladder cancer.
While the studies of ADAMs and their effects on bladder cancer are few in number, several of them can be used for comparing our data. Indeed, van Kampen et al and Jia et al already showed that miR-126 and miR-520f repress the proliferation and metastasis of bladder cancer cells by specific targeting.30,31 Additionally, Fu et al and Fröhlich et al reported that ADAM10 and ADAM12 are involved in regulating the proliferation and invasion process of bladder cancer cells.32,33 Nevertheless, in our study, the expression of a panel of various ADAMs in bladder cancer tissue was investigated for the first time. Our results also showed that the inhibited expression of ADAM15 via miR-3174 significantly reduced the proliferation and metastasis of the patient-derived bladder cancer cells. These results were consistent with previous results from Lorenzatti et al.34 Interestingly, our results show that the expression of ADAM23 in the tumor tissue was lower compared to the non-tumor tissue. Also, the expression of ADAM28 and ADAM33 in the tumor tissue was slightly lower compared to the non-tumor tissue, although no statistical significance was found. Furthermore, the expression of ADAM17, ADAM15, ADAM12, and ADAM10 was significantly higher in the tumor tissue than in the non-tumor tissue. The expression of ADAM9 and ADAM22 was found similar in both the tumor and non-tumor tissue. Moreover, these results were notconsistent with the results from Tyan et al and Yang et al, who reported that ADAM28 is overexpressed in bladder transitional cell carcinoma.35,36 More clinical specimens will be collected for related testing in the future.
The biological behavior of cancer cells in tumor tissues is generally affected by the extracellular matrix (ECM) and the recognition, adhesion, and interaction between cells. In this perspective, the ECM modulates the behavior and features of solid tumors.37,38 Here, the collagen and osteopontin components of the ECM can modify the microenvironment in solid tumors to regulate drug-resistance, proliferation, and metastasis of tumor cells. As a membrane protein, the ADAMs can function as a key regulator of adhesion and interaction between cells in their ECM. Here, ADAM15 promotes the extracellular shedding of E-cadherin, which is involved in cell activation and proliferation. With such an effect, the down-regulation of ADAM15 could kill bladder cancer cells. Thus, the results of the present work also provide a theoretical basis for future targeted treatment of bladder cancer. Moreover, ADAMs also have other functions. For instance, Zhang et al and Dosch et al suggested that ADAM17 and ADAM10 mediate the cleaving and activation of the NOTCH (Notch Signaling pathway) protein and enhance the EMT and resistance of tumor cells.39,40 A such, targeting these ADAMs could be an approach for antitumor strategies. Overall, thanks to the various roles that the ADAMs play in tumor biology, it is valuable to further examine their functions and mechanisms regarding the regulation of bladder cancer in the future.
The interaction between miRNAs/genes plays important roles in the tumorigenesis of human malignancies.41,42 The investigation on genes and miRNAs seems to be a novel diagnostic instrument in the medical field; however, the limit is that often the same gene/miRNA plays a role in different cancer types.43 Besides the miRs, some new future targets of therapy, including the immune system, are the promising strategies for bladder cancer treatment.44–47
Acknowledgments
We thank Dr Yu Cao in the Department of Immunology, H. Lee Moffitt Cancer Center & Research Institute, Tampa, FL, 33612, USA, for his kind advice.
Author Contributions
All authors made substantial contributions to the design and conception, acquisition, analysis, or interpretation of data. All authors took part in either drafting or revising the manuscript. All authors also gave final approval of the version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Kamat AM, Hahn NM, Efstathiou JA, et al. Bladder cancer. Lancet. 2016;388(10061):2796–2810. doi:10.1016/S0140-6736(16)30512-8
2. Babjuk M, Burger M, Compérat EM, et al. European association of urology guidelines on non-muscle-invasive bladder cancer (TaT1 and carcinoma in situ) - 2019 update. Eur Urol. 2019;76(5):639–657. doi:10.1016/j.eururo.2019.08.016
3. Poli G, Brancorsini S, Cochetti G, Barillaro F, Egidi MG, Mearini E. Expression of inflammasome-related genes in bladder cancer and their association with cytokeratin 20 messenger RNA. Urol Oncol. 2015;33(12):
4. Miller MA, Sullivan RJ, Lauffenburger DA. Molecular pathways: receptor ectodomain shedding in treatment, resistance, and monitoring of cancer. Clin Cancer Res. 2017;23(3):623–629. doi:10.1158/1078-0432.CCR-16-0869
5. Rossello A, Nuti E, Ferrini S, Fabbi M. Targeting ADAM17 sheddase activity in cancer. Curr Drug Targets. 2016;17(16):1908–1927. doi:10.2174/1389450117666160727143618
6. Dreymueller D, Ludwig A. Considerations on inhibition approaches for proinflammatory functions of ADAM proteases. Platelets. 2017;28(4):354–361. doi:10.1080/09537104.2016.1203396
7. Sépult C, Bellefroid M, Rocks N, et al. ADAM10 mediates malignant pleural mesothelioma invasiveness. Oncogene. 2019;38(18):3521–3534. doi:10.1038/s41388-018-0669-2
8. Pastorini P, Milano G, Toubol J, Raymond G, Cambon P, Lalanne CM. The diagnostic and prognostic value of urinary polyamine measurement in bladder cancer. Urol Res. 1981;9(1):13–16. doi:10.1007/BF00256832
9. Amendola MA, Glazer GM, Grossman HB, Aisen AM, Francis IR. Staging of bladder carcinoma: MRI-CT-surgical correlation. Am J Roentgenol. 1986;146(6):1179–1183. doi:10.2214/ajr.146.6.1179
10. Prout GR
11. Poplin R, Chang PC, Alexander D, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat Biotechnol. 2018;36(10):983–987. doi:10.1038/nbt.4235
12. Li F, Wei A, Bu L, et al. Procaspase-3-activating compound 1 stabilizes hypoxia-inducible factor 1α and induces DNA damage by sequestering ferrous iron. Cell Death Dis. 2018;9(10):1025. doi:10.1038/s41419-018-1038-3
13. Guan F, Ding R, Zhang Q, et al. WX-132-18B, a novel microtubule inhibitor, exhibits promising anti-tumor effects. Oncotarget. 2017;8(42):71782–71796. doi:10.18632/oncotarget.17710
14. Chen Y, Zeng Q, Liu X, et al. LINE-1 ORF-1p enhances the transcription factor activity of pregnenolone X receptor and promotes sorafenib resistance in hepatocellular carcinoma cells. Cancer Manag Res. 2018;Oct:4421–4438. doi:10.2147/CMAR.S176088
15. Ji Q, Xu X, Li L, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8(10):e3103. doi:10.1038/cddis.2017.499
16. Liang Y, Xu X, Wang T, et al. The EGFR/miR-338-3p/EYA2 axis controls breast tumor growth and lung metastasis. Cell Death Dis. 2017;8(7):e2928. doi:10.1038/cddis.2017.325
17. Li L, Liang Y, Kang L, et al. Transcriptional regulation of the warburg effect in cancer by SIX1. Cancer Cell. 2018;33(3):368–385.e7. doi:10.1016/j.ccell.2018.01.010
18. Shao Z, Li Y, Dai W, et al. ETS-1 induces Sorafenib-resistance in hepatocellular carcinoma cells via regulating transcription factor activity of PXR. Pharmacol Res. 2018;135:188–200. doi:10.1016/j.phrs.2018.08.003
19. Feng F, Jiang Q, Cao S, et al. Pregnane X receptor mediates sorafenib resistance in advanced hepatocellular carcinoma. Biochim Biophys Acta Gen Subj. 2018;1862(4):1017–1030. doi:10.1016/j.bbagen.2018.01.011
20. Wei L, Lun Y, Zhou X, et al. Novel urokinase-plasminogen activator inhibitor SPINK13 inhibits growth and metastasis of hepatocellular carcinoma in vivo. Pharmacol Res. 2019;143:73–85. doi:10.1016/j.phrs.2019.03.009
21. Xu X, Zhang G, He L, Zhu Y. Clinicopathological impacts of c-Met overexpression in bladder cancer: evidence from 1,336 cases. Onco Targets Ther. 2019;12:2695–2702. doi:10.2147/OTT.S197540
22. Dinges SS, Hohm A, Vandergrift LA, et al. Cancer metabolomic markers in urine: evidence, techniques and recommendations. Nat Rev Urol. 2019;16:339–362. doi:10.1038/s41585-019-0185-3
23. Xie Y, Li P, Gao Y, et al. Reduced E-cadherin expression is correlated with poor prognosis in patients with bladder cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(37):62489–62499. doi:10.18632/oncotarget.19934
24. Oo HZ, Seiler R, Black PC, Daugaard M. Post-translational modifications in bladder cancer: expanding the tumor target repertoire. Urol Oncol. 2018;S1078-1439(18)30339-9.
25. Quan J, Pan X, Zhao L, et al. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: a systematic review and meta-analysis. Onco Targets Ther. 2018;4(11):6415–6424. doi:10.2147/OTT.S167853
26. Dong Y, Wu Z, He M, et al. ADAM9 mediates the interleukin-6-induced epithelial-mesenchymal transition and metastasis through ROS production in hepatoma cells. Cancer Lett. 2018;421:1–14. doi:10.1016/j.canlet.2018.02.010
27. Morais M, Dias F, Teixeira AL, Medeiros R. MicroRNAs and altered metabolism of clear cell renal cell carcinoma: potential role as aerobic glycolysis biomarkers. Biochim Biophys Acta Gen Subj. 2017;1861(9):2175–2185. doi:10.1016/j.bbagen.2017.05.028
28. Sun T. Long noncoding RNAs act as regulators of autophagy in cancer. Pharmacol Res. 2018;129:151–155. doi:10.1016/j.phrs.2017.11.009
29. Hasanpourghadi M, Pandurangan AK, Mustafa MR. Modulation of oncogenic transcription factors by bioactive natural products in breast cancer. Pharmacol Res. 2018;128:376–388. doi:10.1016/j.phrs.2017.09.009
30. van Kampen JGM, van Hooij O, Jansen CF, et al. miRNA-520f reverses epithelial-to-mesenchymal transition by targeting ADAM9 and TGFBR2. Cancer Res. 2017;77(8):2008–2017. doi:10.1158/0008-5472.CAN-16-2609
31. Jia AY, Castillo-Martin M, Bonal DM, Sánchez-Carbayo M, Silva JM, Cordon-Cardo C. MicroRNA-126 inhibits invasion in bladder cancer via regulation of ADAM9. Br J Cancer. 2014;110(12):2945–2954. doi:10.1038/bjc.2014.245
32. Fu L, Liu N, Han Y, Xie C, Li Q, Wang E. ADAM10 regulates proliferation, invasion, and chemoresistance of bladder cancer cells. Tumour Biol. 2014;35(9):9263–9268. doi:10.1007/s13277-014-2201-9
33. Fröhlich C, Albrechtsen R, Dyrskjøt L, Rudkjaer L, Ørntoft TF, Wewer UM. Molecular profiling of ADAM12 in human bladder cancer. Clin Cancer Res. 2006;12(24):7359–7368. doi:10.1158/1078-0432.CCR-06-1066
34. Lorenzatti Hiles G, Bucheit A, JR R, et al. ADAM15 is functionally associated with the metastatic progression of human bladder cancer. PLoS One. 2016;11(3):e0150138. doi:10.1371/journal.pone.0150138
35. Tyan YC, Yang MH, Chen SC, et al. Urinary protein profiling by liquid chromatography/tandem mass spectrometry: ADAM28 is overexpressed in bladder transitional cell carcinoma. Rapid Commun Mass Spectrom. 2011;25(19):2851–2862. doi:10.1002/rcm.5169
36. Yang MH, Chu PY, Chen SC, et al. Characterization of ADAM28 as a biomarker of bladder transitional cell carcinomas by urinary proteome analysis. Biochem Biophys Res Commun. 2011;411(4):714–720. doi:10.1016/j.bbrc.2011.07.010
37. Hou J, Hong Z, Feng F, et al. A novel chemotherapeutic sensitivity-testing system based on collagen gel droplet embedded 3D-culture methods for hepatocellular carcinoma. BMC Cancer. 2017;17(1):729. doi:10.1186/s12885-017-3706-6
38. Liu G, Fan X, Tang M, et al. Osteopontin induces autophagy to promote chemo-resistance in human hepatocellular carcinoma cells. Cancer Lett. 2016;383(2):171–182. doi:10.1016/j.canlet.2016.09.033
39. Dosch J, Ziemke E, Wan S, et al. Targeting ADAM17 inhibits human colorectal adenocarcinoma progression and tumor-initiating cell frequency. Oncotarget. 2017;8(39):65090–65099. doi:10.18632/oncotarget.17780
40. Zhang Y, Li D, Jiang Q, et al. Novel ADAM-17 inhibitor ZLDI-8 enhances the in vitro and in vivo chemotherapeutic effects of Sorafenib on hepatocellular carcinoma cells. Cell Death Dis. 2018;9(7):743. doi:10.1038/s41419-018-0804-6
41. Poli G, Egidi MG, Cochetti G, Brancorsini S, Mearini E. Relationship between cellular and exosomal miRNAs targeting NOD-like receptors in bladder cancer: preliminary results. Minerva Urol Nefrol. 2019. doi:10.23736/S0393-2249.19.03297-1
42. Guelfi G, Cochetti G, Stefanetti V, et al. Next generation sequencing of urine exfoliated cells: an approach of prostate cancer microRNAs research. Sci Rep. 2018;8(1):7111. doi:10.1038/s41598-018-24236-y
43. Mezzasoma L, Antognelli C, Del Buono C, et al. Expression and biological-clinical significance of hTR, hTERT and CKS2 in washing fluids of patients with bladder cancer. BMC Urol. 2010;10:17. doi:10.1186/1471-2490-10-17
44. Mearini E, Poli G, Cochetti G, Boni A, Egidi MG, Brancorsini S. Expression of urinary miRNAs targeting NLRs inflammasomes in bladder cancer. Onco Targets Ther. 2017;22(10):2665–2673. doi:10.2147/OTT.S132680
45. Pastina P, Nardone V, Croci S, et al. Anti-cancer activity of dose-fractioned mPE ± bevacizumab regimen is paralleled by immune-modulation in advanced squamous NSLC patients. J Thorac Dis. 2017;Sep(9):3123–3131. doi:10.21037/jtd.2017.08.68
46. Felsenstein KM, Theodorescu D. Precision medicine for urothelial bladder cancer: update on tumour genomics and immunotherapy. Nat Rev Urol. 2018;15(2):92–111. doi:10.1038/nrurol.2017.179
47. Saad FT, Hincal E, Kaymakamzade B. Dynamics of immune checkpoints, immune system, and BCG in the treatment of superficial bladder cancer. Comput Math Methods Med. 2017;2017:3573082. doi:10.1155/2017/3573082
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.