Back to Journals » OncoTargets and Therapy » Volume 11
The methylation profiles of PRDM promoters in non–small cell lung cancer
Authors Tan SX, Hu RC, Xia Q, Tan YL, Liu JJ, Gan GX, Wang LL
Received 11 November 2017
Accepted for publication 25 March 2018
Published 22 May 2018 Volume 2018:11 Pages 2991—3002
DOI https://doi.org/10.2147/OTT.S156775
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Jianmin Xu
Shuang-xiang Tan,1,2 Rui-cheng Hu,1,2 Qian Xia,2 Yong-li Tan,1 Jing-jing Liu,1 Gui-xiang Gan,1 Li-le Wang1
1Hunan Province Institute of Gerontology, Hunan Provincial People’s Hospital/The First Affiliated Hospital of Hunan Normal University, Changsha, China; 2Department of Respiratory Medicine, Hunan Provincial People’s Hospital/The First Affiliated Hospital of Hunan Normal University, Changsha, China
Background: Non–small cell lung cancer (NSCLC) is one of the leading malignant tumors worldwide. Aberrant gene promoter methylation contributes to NSCLC, and PRDM is a tumor suppressor gene family that possesses histone methyltransferase activity. This study aimed to investigate whether aberrant methylation of PRDM promoter is involved in NSCLC.
Materials and methods: Primary tumor tissues, adjacent nontumorous tissues, and distant lung tissues were collected from 75 NSCLC patients including 52 lung squamous cell carcinoma (LSCC) patients and 23 lung adenocarcinoma patients. The expression of PRDMs was detected by polymerase chain reaction (PCR), Western blot, and immunohistochemical analysis. The methylation of PRDM promoters was detected by methylation-specific PCR. The correlation of methylation and expression of PRDMs with clinicopathological characteristics of patients were analyzed.
Results: mRNA expression of PRDM2, PRDM5, and PRDM16 was low or absent in tumor tissues compared to distant lung tissues. The methylation frequencies of PRDM2, PRDM5, and PRDM16 in tumor tissues were significantly higher than those in distal lung tissues. In LSCC patients, methylation of PRDM2 and PRDM16 was correlated with smoking status and methylation of PRDM5 was correlated with tumor differentiation.
Conclusion: The expression of PRDM2, PRDM5, and PRDM16 is low or absent in NSCLC, and this is mainly due to gene promoter methylation. Smoking may be an important cause of PRDM2 and PRDM16 methylation in NSCLC.
Keywords: lung tumor, PR domain zinc finger protein, epigenetics, methylation, gene expression
Introduction
Lung cancer has been a globally important public health problem for decades; it was the seventh leading cause of disease deaths in 1990 and is expected to be the fifth leading cause of disease deaths in 2010.1 Despite encouraging progress on tumor therapy in the past decades, the prognosis of lung cancer has not been markedly improved. Non–small cell lung cancer (NSCLC) accounts for ~80% of lung cancer cases, and its prognosis is relatively better than small cell lung cancer. Epidemiological data show that the average survival of a NSCLC patient is <8 months if left untreated.2 The overall 5-year survival rate for NSCLC is <17% because of the lack of early diagnosis and timely therapy. Cigarette smoking is an important risk factor of lung cancer, and other environmental risk factors for lung cancer include exposure to secondhand tobacco smoking, occupational lung carcinogens, radiation, and air pollution.3
Epigenetic regulation is a process that influences the accessibility of DNA to transcriptional regulatory factors that activate or repress gene expression. Epigenetic mechanisms such as aberrant DNA methylation, histone modification, chromatin remodeling, and functional noncoding RNAs have contributed to the pathogenesis of lung cancer, providing additional markers for early detection, monitoring, prognosis, risk assessment, and personalized treatment of lung cancer.4,5
PR domain zinc finger proteins (PRDMs) are evolutionarily conserved zinc finger transcription factors with tissue-specific expression profile and they play key roles during cell differentiation, organ development, and human diseases.6,7 PRDMs regulate target gene expression through PR domain-dependent histone modification at gene promoters.6 PRDM genes are usually inactivated in hematological malignancies and solid cancers, and the inactivation mechanisms include promoter methylation, homozygous deletions, frameshift mutation, missense mutations, and PR domain deletion.8
Currently, the role of PRDMs in lung cancer remains unclear. PRDM1, PRDM2, PRDM14, and PRDM16 have been implicated in lung cancer pathogenesis, but the results were contradictory.9–14 We hypothesized that methylation-mediated PRDM inactivation may participate in the pathogenesis of NSCLC. In this study, clinical tissue specimens of primary carcinoma tissue, tumor adjacent tissue, and distant lung tissue were collected from 75 NSCLC patients, and the expression and methylation of PRDMs were analyzed. Our results demonstrated that DNA methylation of PRDM2, PRDM5, and PRDM16 was correlated with the malignancy of NSCLC.
Materials and methods
Patients and tissue specimens
A total of 52 lung squamous cell carcinoma (LSCC) patients and 23 lung adenocarcinoma (LAC) patients were enrolled in this study. The clinical characteristics of the patients are shown in Table 1. All diagnoses were confirmed by radiology combined with histopathology, and staging was according to the criteria provided by the International Association for the Study of Lung Cancer in 2009. Cancerous tissue, paracancerous tissue (<2 cm from primary carcinoma), and distant lung tissue (at least 4 cm away from primary carcinoma) were collected from each patient. Tissue specimens were paraformaldehyde-fixed for the preparation of paraffin-embedded sections or frozen in liquid nitrogen for later use. The study protocols have been approved by the ethics committee of Hunan Provincial People’s Hospital, and written informed consent was obtained from all the patients.
  | Table 1 Clinical characteristics of the patients |
Reverse transcription polymerase chain reaction (RT-PCR)
Total RNA was extracted from the tissues by using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA), and cDNA was synthesized by reverse transcription by using RT kit (Promega Corporation, Fitchburg, WI, USA) according to the manufacturer’s instructions. PCR was performed with TaqMasterMix (Promega, Madison, WI, USA). The oligonucleotide sequences of primers were designed with an online software (http://frodo.wi.mit.edu/) and synthesized by Takara. The sequences of primers, annealing temperatures, and the length of products are shown in Table 2. After 35 cycles, PCR products were electrophoresed on 1.5% agarose gel and stained with ethidium bromide, then the images were scanned by using ultraviolet (UV) gel imaging system. The expression level of target gene in each sample was calculated relative to that of β-actin.
  | Table 2 Primers used for reverse transcription polymerase chain reaction |
Methylation-specific PCR (MSP)
Genomic DNA was extracted from the tissues using Universal Genomic DNA Extraction Kit (Takara, Tokyo, Japan), and modified by bisulfite treatment with EZ-DNA methylation kit (Zymo Research, Orange, CA, USA) according to the manufacturer’s instructions, then used for MSP. Primer pairs for methylated and unmethylated target gene were designed with online software (http://www.urogene.org/methprimer/index1.html) and synthesized by Takara (Japan). The sequences of primers, annealing temperatures, and the length of products are shown in Table 3. After 35 cycles, PCR products were electrophoresed on 2% agarose gel and stained with ethidium bromide, then the images were scanned by using UV gel imaging system.
  | Table 3 Primers used for methylation specific-polymerase chain reaction |
Western blot analysis
Tissues were lysed in RIPA lysis buffer, lysate was centrifuged at 15,000× g for 30 minutes at 4°C, the supernatant was collected and protein concentration was measured by BSA method. Fifty microgram protein samples were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). Next, the membranes were blocked with 5% non-fat dry milk for 1 hour at room temperature and then incubated with specific antibody (Abcam, Cambridge, MA, USA) against PRDM2 (1:500 dilution), PRDM5 (1:500 dilution), PRDM16 (1:500 dilution), or β-actin (1:2,000 dilution) at 4°C overnight. The membranes were washed and then incubated with horseradish peroxidase-conjugated secondary antibody for 1 hour at room temperature. Finally, the membranes were developed using ECL kit (Pierce, Rockford, IL, USA) and exposed to X-ray film for analysis by Image.plus5.1 software (Media Cybernetics, Rockville, MD, USA).
Immunohistochemistry (IHC)
IHC was performed on paraformaldehyde-fixed and paraffin-embedded tissue sections. Tissue sections (4 μm thickness) were deparaffinized with xylene and rehydrated with graded alcohol, then treated with antigen retrieval solution (10 mmol/L sodium citrate buffer, pH 6.0). The sections were incubated with specific antibody (Abcam; 1:150 dilution, omitted in negative control) against PRDM2, PRDM5, PRDM16, or β-actin at 4°C overnight, then incubated with biotinylated secondary antibody (1:1,000 dilution), followed by avidin-biotin-peroxidase complex, according to the manufacturer’s instructions. Finally, the sections were incubated with 3′,3′-diaminobenzidine and counterstained with Harris’ modified hematoxylin. The staining intensity was graded by using a 4-point scale: 0, no staining; 1, light yellow; 2, brown; and 3, dark brown. The proportion of cells stained was assessed by using a 3-point scale: 1, <30% staining; 2, 30%–70% staining; and 3, >70% staining. The combined value of 4-point scale and 3-point scale was assessed as follows: 0–1, negative staining; 2, weakly positive staining; 3, positive staining; and ≥4, strong positive staining.
Statistical analysis
Data are presented as mean ± standard deviation. All statistical analyses were performed by using SPSS13.0 software (SPSS Inc., Chicago, IL, USA). Student’s t-test or one-way analysis of variance followed by the Newman–Keuls test was performed to analyze numerical data. Chi-square test was performed to analyze categorical data. P<0.05 was considered significant.
Results
mRNA expression of PRDM family members in lung cancer tissues
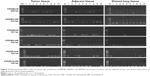
The mRNA expression of PRDM1, PRDM2, PRDM3, PRDM5, PRDM6, PRDM7, PRDM8, PRDM10, PRDM12, PRDM13, PRDM14, PRDM15, PRDM16, and PRDM17 was detected by RT-PCR in primary carcinoma tissues, tumor adjacent tissues, and distant lung tissues. The results showed that mRNA expression levels of PRDM2, PRDM5, and PRDM16 in primary carcinoma tissues were lower than in tumor adjacent tissues and distant lung tissues, mRNA expression levels of PRDM1, PRDM10, PRDM14, and PRDM15 in primary carcinoma tissues were not different from tumor adjacent tissues and distant lung tissues, while mRNA expression level of PRDM6 was higher in primary carcinoma tissues than in tumor adjacent tissues and distant lung tissues (Figures 1 and 2; Tables 4 and 5). For other PRDMs, we could not detect their mRNA expression (data not shown).
Methylation status of the promoters of PRDM family members in lung cancer tissues
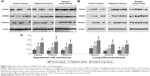
We wondered whether decreased PRDM2, PRDM5, and PRDM16 mRNA expression in tumor tissues was due to promoter methylation; hence, we performed MSP and the results are shown in Figures 3 and 4 and Table 6.
  | Table 6 PRDM2, PRDM5, and PRDM16 methylation status in lung tissues |
In patients with LSCC, PRDM2 gene methylation (with fully methylated and partially methylated) frequency was 67.3%, 50.0%, and 17.3%, respectively, in tumor tissues, adjacent tissues, and distant lung tissues. In patients with LAC, PRDM2 gene methylation frequency was 78.3%, 34.8%, and 21.7%, respectively, in tumor tissues, adjacent tissues, and distant lung tissues. In patients with LSCC, PRDM5 gene methylation was 73.1%, 44.2%, and 21.1%, respectively, in tumor tissues, adjacent tissues, and distant lung tissues. In patients with LAC, PRDM5 gene methylation frequency was 82.6%, 47.8%, and 17.4%, respectively, in tumor tissues, adjacent tissues, and distant lung tissues. In patients with LSCC, PRDM16 gene methylation was 80.8%, 40.4%, and 21.2%, respectively, in tumor tissues, adjacent tissues, and distant lung tissues. In patients with LAC, PRDM16 gene methylation frequency was 82.6%, 52.2%, and 30.4%, respectively, in tumor tissues, adjacent tissues, and distant lung tissues.
Protein expression of PRDM family members in lung cancer tissues
To confirm the results of RT-PCR, we performed Western blot analysis on lung cancer tissues. The results showed that protein expression levels of PRDM2, PRDM5, and PRDM16 in primary carcinoma tissues were lower than in tumor adjacent tissues and distant lung tissues (Figure 5). Furthermore, IHC staining showed that protein expression levels of PRDM2, PRDM5, and PRDM16 in primary carcinoma tissues were lower than in tumor adjacent tissues and distant lung tissues (Figure 6; Table 7).
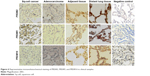  | Figure 6 Representative immunohistochemical staining of PRDM2, PRDM5, and PRDM16 in clinical samples. |
  | Table 7 Immunohistochemical staining of PRDM2, PRDM5, and PRDM16 in clinical specimens |
Correlation of PRDM2, PRDM5, and PRDM16 methylation and clinical aspects of lung cancer patients
Next we analyzed the correlation of PRDM2, PRDM5, and PRDM16 gene methylation status with the age, gender, smoking, tumor differentiation, clinical stage, lymph node metastasis in lung squamous carcinoma, and LAC patients. The results showed that PRDM2 and PRDM16 gene methylation in LSCC patients was related to smoking, and methylation status of PRDM5 gene was associated with tumor differentiation of LSCC (Table 8). However, PRDM2, PRDM5, and PRDM16 gene methylation showed no correlation with age, gender, smoking, tumor differentiation, clinical stage, and lymph node metastasis in LAC patients (Table 9).
  | Table 8 Correlation of PRDM2, PRDM5, and PRDM16 methylation with clinical characteristics of lung squamous cell carcinoma |
  | Table 9 Correlation of PRDM2, PRDM5, and PRDM16 methylation with clinical characteristics of lung adenocarcinoma |
Discussion
In the past few decades, comprehensive utilization of various treatment methods has significantly improved cancer survival, but the prognosis of patients with lung cancer did not significantly change. Tobacco is still the most important risk factor for lung cancer.3 Because environmental tobacco exposure is a risk factor of lung cancer, the role of epigenetic mechanisms in the pathogenesis of lung cancer has attracted more attention. Tobacco exposure is directly related to gene methylation abnormality and the inactivation of tumor suppressor gene expression.15 Thus, abnormal methylation plays an important role in the pathogenesis of lung cancer and provides a reference for early diagnosis and individualized treatment of lung cancer.16
At present, 17 PRDM coding genes from the human genome have been identified and named PRDM1 to PRDM17.6,7 PRDM is widely involved in the pathogenesis of many kinds of tumors and is an important tumor suppressor gene family.8 In this study, we collected tumor tissues and surrounding tissues from patients with NSCLC. RT-PCR, Western blot, and IHC staining showed that PRDM2, PRDM5, PRDM16 mRNA, and protein expression levels of distant lung tissues were lower than in cancer tissues and adjacent tissues. These results suggest that PRDM2, PRDM5, and PRDM16 expression may be suppressed due to gene hypermethylation in lung cancer. Methylation-specific PCR confirmed that PRDM2, PRDM5, and PRDM16 gene methylation frequency of tumor tissues was higher than that of the adjacent tissues.
LSCC is more common in smokers, and abnormal methylation is a common mechanism of smoking-related diseases.15 Our results showed that in LSCC, PRDM2 and PRDM16 gene methylation was correlated with the smoking of the patients, indicating that smoking may be an important cause of PRDM2 and PRDM16 gene methylation in squamous cell carcinoma of the lung. Yoon et al reported that the single-nucleotide polymorphism of PRDM2 gene was associated with the risk of lung cancer.12 However, we failed to find significant correlation between PRDM2 and PRDM16 gene methylation and clinical pathological characteristics of LAC patients. This may be due to the small sample size of this study, but PRDM2 and PRDM16 gene methylation may differ between LAC and squamous cell carcinoma of the lung. Further studies are needed to elucidate the mechanism.
Notably, we found that PRDM5 gene methylation in LSCC was related to tumor differentiation because poorly differentiated squamous cell carcinoma had higher methylation ratio. PRDM5 may be involved in the regulation of differentiation of LSCC and PRDM5 gene methylation may help evaluate the prognosis of squamous cell carcinoma of the lung. The methylation of PRDM5 resulted in decreased expression of PRDM5 in nasopharyngeal carcinoma, esophageal cancer, gastric cancer, cervical cancer, colon cancer, and other tumors.16–19 However, the expression of PRDM5 in lung cancer has not been reported. Our study provided the first evidence that the methylation of PRDM5 gene is involved in the pathogenesis of lung cancer and is related to tumor differentiation in patients with LSCC. PRDM5 lacks intrinsic histone methyltransferase activity, but it can recruit other histone methyltransferases to the promoter regions and regulate the transcription of tumor-associated genes.20 In addition, PRDM5 interacts with insulator factors to modulate chromatin organization and regulate target gene transcription.21 Further mechanistic studies are needed to understand the role of PRDM5 in lung cancer.
PRDM16 participates in the regulation of cell differentiation and tissue and organ development.22 Current studies mainly focus on PRDM16 and blood tumors, and the results showed that PRDM16 promoted the occurrence of bone marrow hyperplasia syndrome and leukemia.23,24 Man et al reported that PRDM16 gene copy number increased in osteosarcoma patients.25 Takahata et al reported that PRDM16 expression increased in gastric cancer cells and promoted tumor development by inhibiting TGF-beta signaling pathway.14 These findings suggest that PRDM16 may act as an oncogene to promote tumor. However, a recent study reported that PRDM16 functions as a tumor suppressor and its highly specific H3 K4 methyltransferase activity is essential for suppressing MLL1-rearranged acute leukemia.26 Currently, the expression of PRDM16 has not been reported in lung cancer patients. Our study is the first to show that PRDM16 expression is repressed in lung cancer patients, which is related to the methylation of PRDM16 promoter. These data suggest that PRDM16 may be a tumor suppressor in lung cancer.
Although we reported that PRDM2, PRDM5, and PRDM16 gene methylation was correlated to their low expression levels in lung cancer samples, we could not exclude other mechanisms such as chromosome translocation, microsatellite instability, allelic loss, gene copy number variation, gene mutation, and non-coding RNAs that contribute to the regulation of PRDM2, PRDM5, and PRDM16 expression in NSCLC. In addition, the role of PRDM2, PRDM5, and PRDM16 in the pathogenesis of NSCLC may be related to the influence of genomic stability due to histone methyltransferase activity of PRDMs. Further studies are needed to provide deep understanding of the role of PRDMs in lung cancer.
In summary, we found that the expression of PRDM2, PRDM5, and PRDM16 is low or absent in NSCLC tissues, and this is mainly due to gene promoter methylation. Smoking may be an important cause of PRDM2 and PRDM16 methylation in NSCLC. While our data indicate that PRDM status may be helpful for the diagnosis of lung cancer, large-scale studies are needed to evaluate the potential of PRDMs as diagnostic and prognostic markers of lung cancer.
Acknowledgments
This study was supported by Hunan Province Natural Science Foundation (No 11JJ3110) and Hunan Provincial People’s Hospital Charity Fund Foundation.
Disclosure
The authors report no conflicts of interest in this work.
References
Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. | ||
Wao H, Mhaskar R, Kumar A, Miladinovic B, Djulbegovic B. Survival of patients with non-small cell lung cancer without treatment: a systematic review and meta-analysis. Syst Rev. 2013;2:10. | ||
Alberg AJ, Brock MV, Ford JG, Samet JM, Spivack SD. Epidemiology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143(5 Suppl):e1S–e29S. | ||
Brzeziańska E, Dutkowska A, Antczak A. The significance of epigenetic alterations in lung carcinogenesis. Mol Biol Rep. 2013;40(1):309–325. | ||
Lu F, Zhang HT. DNA methylation and nonsmall cell lung cancer. Anat Rec (Hoboken). 2011;294(11):1787–1795. | ||
Hohenauer T, Moore AW. The Prdm family: expanding roles in stem cells and development. Development. 2012;139(13):2267–2282. | ||
Fog CK, Galli GG, Lund AH. PRDM proteins: important players in differentiation and disease. Bioessays. 2012;34(1):50–60. | ||
Mzoughi S, Tan YX, Low D, Guccione E. The role of PRDMs in cancer: one family, two sides. Curr Opin Genet Dev. 2016;36:83–91. | ||
Yu Z, Sato S, Trackman PC, Kirsch KH, Sonenshein GE. Blimp1 activation by AP-1 in human lung cancer cells promotes a migratory phenotype and is inhibited by the lysyl oxidase propeptide. PLoS One. 2012;7(3):e33287. | ||
He L, Yu JX, Liu L, et al. RIZ1, but not the alternative RIZ2 product of the same gene, is underexpressed in breast cancer, and forced RIZ1 expression causes G2-M cell cycle arrest and/or apoptosis. Cancer Res. 1998;58(19):4238–4244. | ||
Du Y, Carling T, Fang W, Piao Z, Sheu JC, Huang S. Hypermethylation in human cancers of the RIZ1 tumor suppressor gene, a member of a histone/protein methyltransferase superfamily. Cancer Res. 2001;61(22):8094–8099. | ||
Yoon KA, Park S, Hwangbo B, et al. Genetic polymorphisms in the Rb-binding zinc finger gene RIZ and the risk of lung cancer. Carcinogenesis. 2007;28(9):1971–1977. | ||
Zhang T, Meng L, Dong W, et al. High expression of PRDM14 correlates with cell differentiation and is a novel prognostic marker in resected non-small cell lung cancer. Med Oncol. 2013;30(3):605. | ||
Takahata M, Inoue Y, Tsuda H, et al. SKI and MEL1 cooperate to inhibit transforming growth factor-beta signal in gastric cancer cells. J Biol Chem. 2009;284(5):3334–3344. | ||
Besingi W, Johansson A. Smoke-related DNA methylation changes in the etiology of human disease. Hum Mol Genet. 2014;23(9):2290–2297. | ||
Shu XS, Geng H, Li L, et al. The epigenetic modifier PRDM5 functions as a tumor suppressor through modulating WNT/β-catenin signaling and is frequently silenced in multiple tumors. PLoS One. 2011;6(11):e27346. | ||
Cheng HY, Chen XW, Cheng L, Liu YD, Lou G. DNA methylation and carcinogenesis of PRDM5 in cervical cancer. J Cancer Res Clin Oncol. 2010;136(12):1821–1825. | ||
Watanabe Y, Kim HS, Castoro RJ, et al. Sensitive and specific detection of early gastric cancer with DNA methylation analysis of gastric washes. Gastroenterology. 2009;136(7):2149–2158. | ||
Watanabe Y, Toyota M, Kondo Y, et al. PRDM5 identified as a target of epigenetic silencing in colorectal and gastric cancer. Clin Cancer Res. 2007;13(16):4786–4794. | ||
Duan Z, Person RE, Lee HH, et al. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol Cell Biol. 2007;27(19):6889–6902. | ||
Galli GG, Carrara M, Francavilla C, et al. Genomic and proteomic analyses of Prdm5 reveal interactions with insulator binding proteins in embryonic stem cells. Mol Cell Biol. 2013;33(22):4504–4516. | ||
Aguilo F, Avagyan S, Labar A, et al. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood. 2011;117(19):5057–5066. | ||
Xiao Z, Zhang M, Liu X, Zhang Y, Yang L, Hao Y. MEL1S, not MEL1, is overexpressed in myelodysplastic syndromes patients with t(1;3) (p36;q21). Leuk Res. 2006;30(3):332–334. | ||
Shing DC, Trubia M, Marchesi F, et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J Clin Invest. 2007;117(12):3696–3707. | ||
Man TK, Lu XY, Jaeweon K, et al. Genome-wide array comparative genomic hybridization analysis reveals distinct amplifications in osteosarcoma. BMC Cancer. 2004;4:45. | ||
Zhou B, Wang J, Lee SY, et al. PRDM16 suppresses MLL1r leukemia via intrinsic histone methyltransferase activity. Mol Cell. 2016;62(2):222–236. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.