Back to Journals » Cancer Management and Research » Volume 12
The Long Noncoding RNA Linc01833 Enhances Lung Adenocarcinoma Progression via MiR-519e-3p/S100A4 Axis
Authors Zhang Y , Li W, Lin Z, Hu J, Wang J, Ren Y, Wei BC, Fan Y, Yang Y
Received 31 August 2020
Accepted for publication 1 October 2020
Published 3 November 2020 Volume 2020:12 Pages 11157—11167
DOI https://doi.org/10.2147/CMAR.S279623
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Yuan Zhang,1,* Wenhua Li,1,* Zongxiang Lin,1 Jingfeng Hu,1 Jingpu Wang,1 Yukai Ren,1 BoChong Wei,1 Yuxia Fan,2 Yang Yang1
1Department of Thoracic Surgery, The First Affiliated Hospital, Zhengzhou University, Zhengzhou 450052, People’s Republic of China; 2Department of Thyroid Surgery, The First Affiliated Hospital of Zhengzhou University, Zhengzhou 450052, People’s Republic of China
*These authors contributed equally to this work.
Correspondence: Yuxia Fan
The First Affiliated Hospital of Zhengzhou University, No. 1 East Jianshe Road, Zhengzhou 450052, People’s Republic of China
Email [email protected]
Yang Yang
The First Affiliated Hospital, Zhengzhou University, No. 1 East Jianshe Road, Zhengzhou 450052, People’s Republic of China
Email [email protected]
Introduction: Lung cancer (LC) is among the most prevalent malignancies worldwide, with extremely high morbidity and mortality rates. Mounting evidence has suggested that the abnormally expressed long noncoding RNA (lncRNA) in lung cancer tissues may play vital roles in tumor progression. In the present research, we aimed to examine the functions and underlying mechanism of linc01833 in lung adenocarcinoma (LUAD).
Methods: qRT-PCR was employed to determine transfection efficiency. CCK-8, transwell invasion assay, Western blotting analysis and qRT-PCR were used to detect proliferation as well as migration of different LUAD cell lines, and were also applied to determine the changes during epithelial–mesenchymal transformation (EMT). Afterwards, bioinformatics and dual-luciferase reporter assay were utilized to explore and to identify the potential corresponding targets of linc01833 and miR-519e-3p.
Results: Linc01833 OE can significantly improve proliferation as well as invasion ability of LC cells and promote the EMT process. Dual-luciferase reporter assay demonstrated that linc01833 could directly bind to miR-519e-3p, thereby inhibiting its expression. Further experiments showed that S100A4 was a direct target of miR-519e-3p. Rescue assay demonstrated that linc01833 acted on the miR-519e-3p/S100A4 axis.
Conclusion: We verified the mechanism of linc01833 in promoting infiltration and metastasis in LUAD. To be specific, linc01833 can function as a competitive endogenous RNA (ceRNA) to adsorb miR-519e-3p through a sponge and regulate S100A4 in lung cancer, thereby being involved in LUAD progression. Collectively, our research provides new insights towards the in-depth understanding of LC progression mechanisms.
Keywords: lung cancer, linc01833, miR-519e-3p, S100A4, EMT
Introduction
Lung cancer (LC) is among the most prevalent malignancies globally due to its high morbidity and mortality rates. Among all types of LC, the incidence of lung adenocarcinoma (LUAD) has exceeded that of lung squamous cell carcinoma (LUSC), showing an increasing trend year by year. Up to now, LUAD accounts for nearly 40–50% of all types of LC. Due to the aggressiveness and metastasis of malignant cancer cells, poor prognosis is common among patients.1 The 5-year survival rate of LC patients is only about 18%, without any significant breakthrough or improvement.2
Mounting attention has been paid to long noncoding RNA (lncRNA) in tumor research due to the accumulative evidence indicating the critical involvement of lncRNA in tumor development. LncRNA can regulate the pathophysiological process of a tumor through various pathways. At present, evidence shows the abnormal expression of lncRNA in diverse types of tumor tissues, with significant effects on tumor proliferation and metastasis. For instance, lncRNA MALATl can enhance the proliferation and metastasis of epithelial ovarian cancer cells,3 but it suppresses breast cancer development.4 Additionally, lncRNA FAL1 can accelerate the proliferation and metastasis of liver cancer cells by functioning as a ceRNA through competitive binding with miR-1236,5 which also promotes progression of colorectal cancer (CRC) by modulating the miR-637/NUPR1 pathway.6 LncRNAMEG3 is capable of suppressing proliferation and invasion of gastric cancer (GC) cells through the p53 signaling pathway, which is considered as a novel therapeutic target for GC.7 Similarly, lncRNAMEG3 can also inhibit metastasis as well as invasion in GC by regulating RNA-21, bringing new hope of long-term survival for GC patients.8 Despite a further understanding of the functional mechanisms of lncRNAs by conducting extensive studies on different lncRNAs, some lncRNAs have not been explored, therefore, their roles in tumorigenesis and tumor development remain unknown. Of note, linc01833 is a target that deserves further study. Our previous research showed that linc01833 was highly expressed in LUAD tissues, which enhanced metastasis and invasion of LC. Nevertheless, the functions of linc01833 in LUAD and its related mechanism have not been extensively reported.
The present study attempted to determine the impact of linc01833 on LUAD progression and to explore its underlying mechanism. For the first time, our experimental results confirmed that linc01833 was capable of enhancing proliferation and invasion in LUAD via the modulation of miR-519e-3p/S100A4, thereby being involved in LUAD tumorigenesis and progression.
Materials and Methods
Data Collection and Bioinformatics Analysis
The Cancer Genome Atlas (TCGA) database was used to extract relevant data for cluster analysis. Linc01833 expression was assessed in non-small cell lung cancer (NSCLC) and normal tissue, followed by construction of a boxplot for intuitive observation and comparison. Afterwards, we analyzed and compared the overall survival (OS) of LC patients with different expression levels of linc01833.
Cell Culture
NSCLC cell lines, A549 and HCC4006, were obtained from Shanghai Cell Bank of Chinese Academy of Sciences, which were maintained in DMEM (Invitrogen, Carlsbad, CA), containing 10% FBS (Gibco, MA, USA) and 100 μg/mL penicillin (Invitrogen) at 37 °C in a humidified incubator with 5% CO2.
Transfection
The linc01833 overexpression (OE) vector, S100A4 OE vector and universal negative control (NC) were provided by Genepharma (Shanghai, China). The 3ʹ-untranslated regions (UTR) in the recognition and binding sites of wild type (WT) or mutant type (mut) of miR-519e-3p in S100A4 were amplified, followed by cloning into pGL3 vector to form pGL3-WT-S100A4-3ʹ-UTR or pGL3-mut-S100A4-3ʹ-UTR. Lipofectamine 2000 (Invitrogen) was adopted for cell transfection, followed by further experiments after incubation.
Cell Proliferation Assay
We measured and evaluated the cell proliferation at different time points using CCK-8 kit. Specifically, all transfected cells were inoculated in a 96-well plate, followed by addition of CCK-8 reagent every 24 h to ensure that CCK-8 was always maintained at a stable target concentration. After continuous incubation of cells in standard environment, the optical density values at 450 nm wavelength were measured at 0, 24, 48, 72, and 96 h by spectrophotometer, followed by measurement of cell proliferation rate. The assays were conducted in triplicate and repeated three times or more to ensure the reliability of results.
RNA Extraction as Well as qRT-PCR
TRIzol reagent (Invitrogen) was utilized to isolate total RNA from NSCLC cells, which was transcribed into cDNA by Prime Script RT kit (Takara, Osaka, Japan), followed by cDNA amplification by qRT-PCR utilizing SYBR Green master mix (Thermo Fisher). Relative expression of mRNA or lncRNA was calculated using the 2−ΔΔCT method, followed by normalization to β-actin levels. Each sample was divided into three groups and analyzed repeatedly.
Transwell Invasion Assay
QCM-EC Matrix cell invasion test (Merck, Darmstadt, Germany) was employed to determine cell invasion capacity. Specifically, the transfected cells suspended in FBS-free medium were added into matrix gel (BD Biosciences, San Jose, CA) in the chamber. Complete medium was further supplemented to lower chambers. Cells were incubated in standard conditions until they crossed the ECM layer and adhered firmly to the base of the fixed membrane. The cells crossing the fixed membrane were fixed with methanol, subjected to H & E staining, observed and counted under microscope at 10× magnification at three randomly selected views. The assay was repeated three times.
Dual-Luciferase Reporter (DLR) Assay
In this study, the dual-luciferase detection kit (Promega, Madison, USA) was used for relevant experimental studies and to analyze the dual-luciferase reporter activity. Specifically, A549 and HCC4006 cells were co-transfected with linc01833 WT, linc01833 mut, miR-519e-3p, S100A4 WT, S100A4 mut, or NC siRNA. Afterwards, cells were digested, dissolved, and centrifuged to collect supernatant, followed by measurement of renilla and firefly luciferase activity by microplate photometer (BioTeck, Winooski, VT, USA). All the experiments were performed in triplicate, followed by calculation of the average value to ensure the reliability of the experimental results.
Western Blot (WB) Analysis
Total protein was extracted from A549 and HCC4006 cells, lysed in RIPA buffer (Roche, Branchburg, NJ) containing protease inhibitor. Protein sample was subjected to SDS-PAGE, transferred to PVDF membrane, blocked with 5% BSA, rinsed with PBS, followed by reaction with primary antibody (anti- E-cadherin, anti-cytokeratin, anti-vimentin, anti-S100A4 and anti-GAPDH) overnight at 4°C. After rewashing, membranes were reacted with HRP-conjugated secondary antibody. Immunoreactivity was observed using Odyssey machine (Roche Diagnostics) with ECL detection reagent (Amersham Biosciences). GAPDH was used as an internal control.
Immunofluorescence Analysis
The slides were soaked and washed three times with phosphate buffer saline (PBS). After fixing with 4% paraformaldehyde for 15 min, the slides were soaked with PBS again. After draining PBS with absorbent paper, the slides were sealed with 5% goat serum at room temperature for 30 min. The slides were incubated with primary antibodies against cytokeratin (1:400 dilution) and vimentin (1:200 dilution) at 4°C overnight, followed by incubation with anti-rabbit secondary antibodies at room temperature for 30 min. Finally, the liquid on the slide was dried using absorbent paper and the slides were sealed with the sealing liquid. The slides were observed and photographed under a fluorescence microscope.
Data Analysis
SPSS version 21.0 (SPSS, Chicago, IL) was used for data statistics and analysis. In brief, categorical data were displayed by frequency and percentage, and continuous data were shown as means ± SD. One-way ANOVA or Student’s t-test was utilized for statistical analysis if appropriate. Pearson correlation was used to analyze the correlation between different gene expressions. Kaplan-Meier method and Log rank test were used to analyze the survival curve. P<0.05 suggested statistical significance.
Results
Biological Characteristics of Linc01833 and Its Relationship with Clinical Features
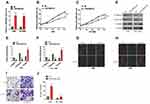
The purpose of this experiment was to examine the biological effects and mechanisms of linc01833 in LUAD. Further mining of the TCGA database uncovered increased linc01833 expression in LUAD tissues (Figure 1A). Survival analysis demonstrated an association of linc01833 expression with OS in LUAD patients (Figure 1B). qRT-PCR assay further revealed significantly higher expression of linc01833 in LUAD tissue than the adjacent normal tissue (Figure 1C).
Biological Role of Linc01833 in LUAD
To confirmthe role of linc01833 in LUAD, linc01833 was overexpressed in two LUAD cell lines A549 and HCC4006, followed by measurement of the transfection efficiency (Figure 2A). CCK-8 assay revealed that linc01833 OE could significantly promote LUAD cell proliferation (Figure 2B and C). In consideration of the important role of EMT in cell metastasis, EMT markers were examined at mRNA and protein levels. WB showed that after OE of linc01833, the expression of cytokeratin and E-cadherin was significantly downregulated in A549 and HCC4006 cells, while that of vimentin was significantly upregulated, suggesting that OE of linc01833 promoted the EMT process of LUAD cells (Figure 2D). Similarly, qRT-PCR analysis showed consistent outcomes in A549 and HCC4006 cell lines (Figure 2E and F). Immunofluorescence subsequently showed significant downregulation of cytokeratin expression but significant upregulation of vimentin expression (Figure 2G and H), which was consistent with the above results. The migration ability of LUAD cells was assessed by transwell invasion assays, showing that OE of linc01833 could significantly improve the migration ability of LUAD (Figure 2I and J). Our experiments confirmed that linc01833 can significantly enhance proliferation, migration and metastasis of LUAD cells, which was also involved inthe EMT process of LUAD cells.
MiR-519e-3p is a Direct Target of Linc01833
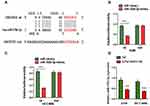
Bioinformatics software was applied to predict and analyze the possible binding miRNA targets of linc01833, aiming to explore its potential mechanism. This showed that miR-519e-3p had the potential to bind with linc01833, and its binding sequence was shown (Figure 3A). Subsequently, we used the DLR assay to determine whether miR-519e-3p could directly bind to linc01833 in A549 and HCC4006 cell lines. Consequently, miR-519e-3p can directly bind with wtlinc01833, while there was insignificant difference in mutlinc01833 group (Figure 3B and C). Moreover, miR-519e-3p expression was significantly inhibited in linc01833 OE group, indicating that linc01833 could negatively modulate miR-519e-3p level (Figure 3D).
Linc01833 Promotes LUAD via Negative Modulation of MiR-519e-3p
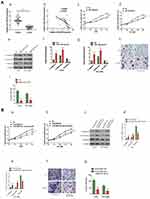
qRT-PCR analysis revealed that miR-519e-3p showed a significantly lower expression trend in LUAD tissue in comparison with adjacent tissue (control) (Figure 4A-a). Increased linc01833 expression in LUAD tissue is generally accompanied by decreased miR-519e-3p expression, and vice versa (Figure 4A-b). In addition, CCK-8 assay demonstrated that miR-519e-3p OE was capable of suppressing LUAD cell proliferation (Figure 4A-c-d). The functions of miR-519e-3p in LUAD were tested by WB (Figure 4A-e) and qRT-PCR (Figure 4A-f-g). The results showed that miR-519e-3p OE led to significantly increased expression of cytokeratin and E-cadherin, but significantly decreased expression of vimentin in LUAD cells, indicating that miR-519e-3p OE was capable of inhibiting the EMT process in LUAD cells (Figure 4A-h-i). Therefore, we believed that miR-519e-3p exerted an inhibitory effect on the progression of LC. To further understand the correlation of linc01833 with miR-519e-3p, reverse assays were conducted. MiR-519e-3p OE could reverse the proliferative effect of linc01833 OE on A549 and HCC4006 cells (Figure 4B-a-b). Compared with linc01833 OE group, miR-519e-3p OE can upregulate the levels of cytokeratin and E-cadherin in LUAD cells, while it can downregulate the expression of vimentin (Figure 4B-c-e). Transwell invasion assay showed similar results (Figure 4B-f-g). The reverse assays revealed that miR-519e-3p OE reversed the effect of linc01833 on proliferation, migration and metastasis of LUAD cells. Therefore, our results suggested that linc01833 promoted LUAD development via negative modulation of miR-519e-3p.
The S100A4 is a Direct Target of MiR-519e-3p
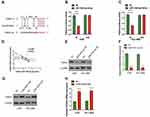
For further investigation of the mechanism underlying the roles of miR-519e-3p on the biological behavior in LUAD, bioinformatics software was applied for prediction and analysis. A potential binding sequence of S100A4 and miR-519e-3p was found (Figure 5A). S100A4 might be a possible target of miR-519e-3p, which was further confirmed by DLR assay (Figure 5B and C). Further investigation found a negative correlation of miR-519e-3p expression with S100A4 expression in LC tissues (Figure 5D). WB and qRT-PCR analysis demonstrated that miR-519e-3p OE could significantly downregulate S100A4 expression, indicating the negative regulatory effects of miR-519e-3p on S100A4 expression (Figure 5E and F). Linc01833 OE can significantly increase the expression of S100A4, either at protein or mRNA level, suggesting that linc01833 can positively regulate the expression of S100A4 (Figure 5G and H).
The Biological Role of Linc01833/MiR-519e-3p/S100A4 Axis in LUAD
Rescue assays were performed to examine the functions of the linc01833/miR-519e-3p/S100A4 axis in invasion and metastasis of LUAD. Compared with the co-transfection group, OE of S100A4 can reverse the effects of linc01833 and miR-519e-3p on LUAD cell proliferation (Figure 6A and B). We also detected the decreased expression of cytokeratin and E-cadherin, and increased vimentin level (Figure 6C–E). These outcomes collectively suggested that S100A4 OE can reverse the effects of OE of linc01833 and miR-519e-3p on EMT and proliferation in LUAD cells.
Discussion
An estimated 1.59 million people die of LC globally every year. Moreover, both incidence and mortality of LC are the highest among all types of malignant tumors, thus attracting more and more attention.9,10 LC is mostly characterized by relatively insidious onset and relatively rapid progression. Despite continuous advances in therapeutic concepts and technology, the 5-year survival rate of LC patients is still not high due to invasion and metastasis of LC. Thus, related research on the progress of LC metastasis has always been a hot spot.
LncRNA generally contains over 200 nucleotides (nt), generally without protein-coding function. LncRNAs can be classified according to different structures, including SenselncRNA, IntroniclncRNA, BidirectionallncRNA, AntisenselncRNA, Intergenic lncRNA (1incRNA), etc.11,12 The expression levels of some lncRNAs can be significantly changed during the development of malignant tumors, which are closely associated with diverse biological processes and may participate in carcinogenesis and tumor development. LncRNAs are likely to be used as diagnostic and therapeutic markers in tumors.13 For instance, lncRNABORG can mediate the relapse and metastasis of breast cancer.14 By regulating SATB2, LncRNA SATB2-AS can affect the microenvironment of tumor immune cell growth and assist to decrease invasion and metastasis of CRC.15 However, there have been no relevant studies on linc01833 so far, thus, we explored its effect and mechanism in LC in our study.
Unlike lncRNA, miRNAs (with 18–25 nt in length) can bind with and degrade 3ʹUTR of target mRNA. Additionally, miRNA also has the ability to regulate coding and noncoding transcriptomes. Therefore, investigation into the regulatory mechanism of the lncRNA-miRNA-mRNA network can provide more comprehensive and in-depth understanding of pathogenesis as well as metastatic mechanisms of tumors, aiming to find possible therapeutic targets. At present, such networks have been widely applied to examine multiple kinds of malignancies.16–18 For instance, lncRNA H1 can function as a competitive ceRNA for modulating EMT and metastasis of bladder cancer via regulating miR-29b-3p expression.19 LncRANSNHG7 upregulates GALNT1 through sponge adsorption of miR-216b to enhance proliferation and liver metastasis in CRC.20 In this study, bioinformatics was applied to reveal that miR-519e-3p is a possible target of linc01833. Combined with software prediction analysis, S100A4 is revealed as a possible target of miR-519e-3p. Afterwards, these results were also verified by DLR assay, showing a direct correlation between linc01833/miR-519e-3p/S100A4. S100A4 is a vital member of the calcium-binding protein S100 family, comprising 101 amino acids. Current studies have shown that S100A4 is involved in diverse physiological activities, including cell movement and adhesion, extracellular matrix reconstruction, tumor angiogenesis, etc.21–24 Meanwhile, studies have found that it is closely related to many malignant tumors, such as pancreatic cancer, CRC, bladder cancer, breast cancer, GC, prostate cancer, nd is significantly expressed in these malignant tumors. Additionally, S100A4 is capable of modulating apoptosis and proliferation, promoting the EMT process of tumor cells, which also has a close association with invasion, lymph node involvement, and distant metastasis of various tumors (including NSCLC).25–34 In this study, we constructed the corresponding relationship of linc01833/miR-519e-3p/S100A4, shedding new light on the mechanism of LUAD progression.
Herein, we revealed the high expression of linc01833 in LUAD tissue and for the first time demonstrated that OE of linc01833 could significantly promote proliferation and invasion in LUAD cells. WB and qRT-PCR analysis were used to detect EMT-related factors, showing that OE of linc01833 also significantly enhanced the EMT process of LC cells. In addition, linc01833 OE can significantly inhibit miR-519e-3p expression. Combining the outcomes from DLR assay, linc01833 was likely to inhibit the expression of miR-519e-3p by sponge adsorption of ceRNA. We also verified that miR-519e-3p OE can inhibit proliferation and the EMT process of LC cells, and can also inhibit S100A4 expression. However, the reverse assays suggest that miR-519e-3p can partially reverse the biological effects of OE of linc01833. OE of S100A4 can significantly enhance proliferation and invasion capacity of LUAD cells and promote their EMT process, which is consistent with the existing literature on the biological role of S100A4.30,34 It can also reverse the combined effect of both OE of Linc01833 and miR-519e-3p on LC cells.
Conclusion
In conclusion, we demonstrated that lncRNA linc01833 could attenuate its negative regulation of S100A4 and promote the infiltration and metastasis of LUAD through sponge adsorption of miR-519e-3p. Meanwhile, miR-519e-3p can inhibit the progression of LUAD through targeted inhibition of S100A4. Taken together, our study has supplemented understanding of the network mechanism of LUAD. However, further investigations are required on the interaction of linc01833/miR-519e-3p/S100A4 axis in the biological network of LC.
Data Sharing Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the manuscript or revising it critically for important intellectual content; agreed manuscript submission to the Journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work. Yuan Zhang and Wenhua Li should be considered joint first author. Yuxia Fan, PhD and Yang Yang, PhD should be considered joint corresponding author.
Funding
Funding information is not applicable.
Disclosure
The authors declare no conflicts of interest or financial ties to disclose.
References
1. Duma N, Santana-Davila R, Molina JR. Non–Small Cell Lung Cancer: epidemiology, Screening, Diagnosis, and Treatment. Mayo Clinic Proce. 2019;94(8):1623–1640. doi:10.1016/j.mayocp.2019.01.013
2. Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA2014;64:9–29.
3. Jin Y, Feng SJ, Qiu S, Shao N, Zheng JH. LncRNA MALAT1 promotes proliferation and metastasis in epithelial ovarian cancer via the PI3K-AKT pathway. Eur Rev Med Pharm Sci. 2017;21:3176–3184.
4. Kim J, Piao HL, Kim BJ, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genetics. 2018;50:1705–1715.
5. Li B, Mao R, Liu C, Zhang W, Tang Y, Guo Z. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of mir-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
6. Wang L, Jiang F, Xia X, Zhang B. LncRNA FAL1 promotes carcinogenesis by regulation of mir-637/NUPR1 pathway in colorectal cancer. Int J Biochem Cell Biol. 2019;106:46–56.
7. Wei GH, Wang X. LncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharm Sci. 2017;21:3850–3856.
8. Dan J, Wang J, Wang Y, et al. LncRNA-MEG3 inhibits proliferation and metastasis by regulating miRNA-21 in gastric cancer. Biomed Pharm. 2018;99:931–938.
9. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA. 2017;67:7–30.
10. Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19.
11. Xu Y, Li J, Wang P, Zhang Z, Wang X. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF-κB. J Cell Biochem. 2019;120(12):19415–19421. doi:10.1002/jcb.29119
12. Li DY, Chen WJ, Luo L, et al. Prospective lncRNA-miRNA-mRNA regulatory network of long non-coding RNA linc00968 in non-small cell lung cancer a549 cells: A miRNA microarray and bioinformatics investigation. Int J Mol Med. 2017;40:1895–1906.
13. Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667.
14. Gooding AJ, Zhang B, Jahanbani FK, et al. The lncRNA BORG drives breast cancer metastasis and disease recurrence. Sci Rep. 2017;7(1):12698. doi:10.1038/s41598-017-12716-6
15. Xu M, Xu X, Pan B, et al. LncRNA SATB2-AS1 inhibits tumor metastasis and affects the tumor immune cell microenvironment in colorectal cancer by regulating SATB2. Mol Cancer. 2019;18(1):135. doi:10.1186/s12943-019-1063-6
16. Wang X, Yin H, Zhang L, et al. The construction and analysis of the aberrant lncRNA-miRNA-mRNA network in non-small cell lung cancer. J Thoracic Dis. 2019;11(5):1772–1778. doi:10.21037/jtd.2019.05.69
17. Wang W, Lou W, Ding B, et al. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging. 2019;11(9):2610–2627. doi:10.18632/aging.101933
18. Zhang G, Pian C, Chen Z, et al. Identification of cancer-related miRNA-lncRNA biomarkers using a basic miRNA-lncRNA network. PLoS One. 2018;13(5):e0196681. doi:10.1371/journal.pone.0196681
19. Lv M, Zhong Z, Huang M, Tian Q, Jiang R, Chen J. LncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by mir-29b-3p as competing endogenous RNA. Biochimica et biophysica acta. Mol Cell Res. 2017;1864:1887–1899.
20. Shan Y, Ma J, Pan Y, Hu J, Liu B, Jia L. LncRNA SNHG7 sponges mir-216b to promote proliferation and liver metastasis of colorectal cancer through upregulating GALNT1. Cell Death Dis. 2018;9(7):722. doi:10.1038/s41419-018-0759-7
21. Sack U, Walther W, Scudiero D, et al. S100A4-induced cell motility and metastasis is restricted by the Wnt/β-catenin pathway inhibitor calcimycin in colon cancer cells. Mol Biol Cell. 2011;22(18):3344–3354. doi:10.1091/mbc.e10-09-0739
22. Keirsebilck A, Bonne S, Bruyneel E, et al. E-cadherin and metastasin (mts-1/S100A4) expression levels are inversely regulated in two tumor cell families. Cancer Res. 1998;58:4587–4591.
23. Boye K, Maelandsmo GM. S100a4 and metastasis: A small actor playing many roles. Am J Pathology. 2010;176(2):528–535. doi:10.2353/ajpath.2010.090526
24. Ambartsumian N, Klingelhofer J, Grigorian M, et al. The metastasis-associated mts1(s100a4) protein could act as an angiogenic factor. Oncogene. 2001;20(34):4685–4695. doi:10.1038/sj.onc.1204636
25. Davies BR, O’Donnell M, Durkan GC, et al. Expression of s100a4 protein is associated with metastasis and reduced survival in human bladder cancer. J Pathology. 2002;196(3):292–299. doi:10.1002/path.1051
26. Egeland EV, Boye K, Park D, et al. Prognostic significance of s100a4-expression and subcellular localization in early-stage breast cancer. Breast Cancer Res Treatment. 2017;162(1):127–137. doi:10.1007/s10549-016-4096-1
27. Boye K, Nesland JM, Sandstad B, Maelandsmo GM, Flatmark K. Nuclear s100a4 is a novel prognostic marker in colorectal cancer. Eur J Cancer. 2010;46(16):2919–2925. doi:10.1016/j.ejca.2010.07.013
28. Shen W, Tan X, Hao F. S100a4 expression is associated with poor prognosis in patients with resectable gastrointestinal stromal tumor. Libyan J Med. 2019;14(1):1659669. doi:10.1080/19932820.2019.1659669
29. Stein U, Arlt F, Walther W, et al. The Metastasis-Associated Gene S100A4 Is a Novel Target of β-catenin/T-cell Factor Signaling in Colon Cancer. Gastroenterology. 2006;131(5):1486–1500. doi:10.1053/j.gastro.2006.08.041
30. Liu L, Qi L, Knifley T, et al. S100A4 alters metabolism and promotes invasion of lung cancer cells by up-regulating mitochondrial complex I protein NDUFS2. J Biol Chem. 2019;294(18):7516–7527. doi:10.1074/jbc.RA118.004365
31. Huang S, Zheng J, Huang Y, et al. Impact of s100a4 expression on clinicopathological characteristics and prognosis in pancreatic cancer: A meta-analysis. Disease Markers. 2016;2016:8137378. doi:10.1155/2016/8137378
32. Ruma IMW, Kinoshita R, Tomonobu N, et al. Embigin promotes prostate cancer progression by S100A4-dependent and-independent mechanisms. Cancers. 2018;10.
33. Tochimoto M, Oguri Y, Hashimura M, et al. S100a4/non-muscle myosin ii signaling regulates epithelial-mesenchymal transition and stemness in uterine carcinosarcoma. Laboratory investigation. J Tech Methods Pathology. 2019;2:123.
34. Chen N, Sato D, Saiki Y, Sunamura M, Fukushige S, Horii A. S100A4 is frequently overexpressed in lung cancer cells and promotes cell growth and cell motility. Biochem Biophysical Res Commun. 2014;447:459–464.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.