Back to Journals » Cancer Management and Research » Volume 11
The Long Noncoding RNA LINC00908 Facilitates Hepatocellular Carcinoma Progression Via Interaction With Sox-4
Received 23 May 2019
Accepted for publication 7 August 2019
Published 30 September 2019 Volume 2019:11 Pages 8789—8797
DOI https://doi.org/10.2147/CMAR.S216774
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Eileen O'Reilly
This paper has been retracted.
Xinhua Hu, Qingxiang Li, Jinfeng Zhang
Department of Laboratory Medicine, Juxian Hospital of Traditional Chinese Medicine, Rizhao 276500, Shandong, People’s Republic of China
Correspondence: Jinfeng Zhang
Department of Laboratory Medicine, Juxian Hospital of Traditional Chinese Medicine, No. 338, South Chengyang Road, Rizhao 276500, Rizhao, People’s Republic of China
Tel +86 633 6887275
Fax +86 633 6887275
Email [email protected]
Background: The hepatocellular carcinoma (HCC) is a highly aggressive and common malignancy worldwide. Accumulating evidence has demonstrated a pivotal role of long noncoding RNAs (lncRNAs) in various tumors. However, the function of intergenic lncRNA LINC00908 is still unknown in HCC.
Methods: The RT-qPCR method was used to quantify the expression of LINC00908. Migration and viability assay were performed to evaluate the in vitro effect and xenograft tumor model was used to measure the in vivo effect. Immunoblot was used to identify the association of LINC00908 with Sox-4 and the stability of Sox-4.
Results: We found a novel lncRNA related to HCC. LINC00908 is highly expressed in tumorous tissues and cell lines compared with normal ones. High LINC00908 expression correlated with advanced TNM stages, tumor size and metastasis. LINC00908 promoted the migration and viability of HCC cells. The in vivo xenograft tumor growth and proliferation were also enhanced by LINC00908 overexpression and inhibited by LINC00908 silence. LINC00908 physically interacted with Sox-4, and the association between LINC00908 and Sox-4 increased the stability of Sox-4 by reducing proteasomal degradation.
Conclusion: Taken together, our current work has identified a novel lncRNA LINC00908 which is critically involved in HCC progression. The LINC00908-Sox-4 axis might provide a new and potential target for pharmaceutical therapies.
Keywords: lncRNA, LINC00908, Sox-4, HCC
Introduction
The hepatocellular carcinoma (HCC) belongs to one of the most aggressive and common types of cancer worldwide.1 HCC patients suffer from a poor prognosis and relatively high recurrence owing to intrahepatic and distal metastasis.2 The mortality rate of HCC in China is highest across the world.3 Despite rapid advances in medical strategies such as surgery, radiotherapy and chemotherapy, there is still no significant improvement for therapy and HCC patients suffer from extremely low 5-year survival.4 Therefore, identifying novel and efficient therapeutic targets is strongly needed for pharmaceutical intervention for HCC.
The long noncoding RNAs (lncRNAs) are designated as the RNA molecules with >200 nucleotides in length with no or minimal protein-coding capacity.5 Recent data have repeatedly demonstrated the pivotal roles of lncRNAs in various biological processes, such as immune regulation, differentiation and apoptosis.6,7 lncRNAs can function through cis- or trans-regulation.8 Furthermore, lncRNAs can also interact with protein or mRNA molecules to affect their stabilities.9,10 Importantly, lncRNAs can actively participate in the tumorigenesis. For example, HNF1A-AS1 represses the malignant phenotypes of HCC cells through direct interaction with SHP1 and functions as a phosphatase activator.11 Instead, HOXA11-AS can serve as a competing endogenous RNA (ceRNA) and facilitate hepatocellular carcinoma development by sponging miR-214-3p.12 Recently, Song et al showed that LINC00908, together with AC064875.2, HOTAIRM1 and RP11-84A19.3 may serve as diagnostic markers for glioma.13 However, the role of LINC00908 in HCC is still elusive.
Here we reported a novel intergenic lncRNA named LINC00908 implicated in HCC progression. LINC00908 is frequently upregulated in HCC tissues and cell lines. Furthermore, overexpression of LINC00908 could promote the viability and migration of HCC cells in vitro and increase xenograft tumor growth in vivo. Mechanistic studies identified that LINC00908 exerted its role by interacting with transcription factor Sox-4. The association between LINC00908 and Sox-4 substantially increased the stability of Sox-4 by attenuating ubiquitin-mediated proteasome degradation of Sox-4. Collectively, we proved that LINC00908 could play an oncogenic role in HCC by stabilizing Sox-4 and therefore offering a novel potential target for depleting liver cancer cells.
Materials And Methods
Cell Culture And Reagents
The 7721, 97L, Huh7, HepG2, Hep3B and the normal L02 cell lines were purchased from Shanghai Cell Biology Institute at Chinese Academy of Sciences. Dulbecco’s modified Eagle’s medium (DMEM) with 9% fetal bovine serum (FBS, Sigma, Shanghai, China) and 150 μg/mL streptomycin (Sigma, Shanghai, China) was used for cell culture with an atmosphere of 5% CO2 in an incubator at 37°C. For reagent details, see Table S1.
Profiling For lncRNAs
Briefly, total RNAs were extracted using the TRIzol Plus RNA Purification Kit (Invitrogen, Shanghai, China). Sequencing for samples was performed at the Beijing Genomics Institute. GeneSpring GX v11.5 was used for background subtraction of the array data. Genes with fold change (FC)>2 and P <0.05 were identified as differentially expressed ones.
Lentiviral Construction
The LINC00908 or Sox-4 sequences were first amplified and cloned into a pWPXL vector (abbreviated as LINC00908 or Sox-4, respectively). The lentiviral constructs were cloned and purchased from GenePharma (Shanghai, China). An empty lentiviral vector was used as overexpression control (designated as “control”). The short hairpin RNA (shRNA) targeting LINC00908 and Sox-4 were designed by GenePharma (Shanghai, China). A scramble (nontarget) RNA was also generated as the corresponding control. Transfection was performed with Lipofectamine 2000 system (Invitrogen, Shanghai, China). Transfection was done at the presence of 1 μg/mL polybrene (Sigma, Shanghai, China).
Human Samples
The HCC samples were surgical archives at Juxian Hospital of Traditional Chinese Medicine from September 2016 to August 2018. Informed and written consent was obtained from all enrolled patients. After treatment by liquid nitrogen, all HCC samples were finally stored at an −80°C refrigerator. Experimental procedures related to human samples were formally approved by Human Research Ethics Committee (HREC) at Juxian Hospital of Traditional Chinese Medicine.
Cytoplasmic And Nuclear Isolation
Thermo Fisher BioReagents (Thermo Fisher Scientific) were used for nucleocytoplasmic separation according to the manufacturer’s protocols. Then, qRT-PCR was used to amplify the results. Specific primers were shown in Table S1.
Migration Assay
Transwell migration assay was conducted using Transwell chemotaxis 24-well chamber (BD Biosciences). ~1×105 cells were placed in the upper chamber with noncoated membranes. After incubation by 24 hrs, cells which migrated into the lower chamber were fixed in paraformaldehyde (4%, Sigma, Shanghai, China) and stained with crystal violet (0.3%, Sigma, Shanghai, China). An inverted microscope (Olympus, Shanghai, China) was used to visualize the results.
In Vivo Tumorigenesis
HepG2 cells transfected with genetically silencing or overexpressing vectors were cultured in DMEM for 24 hrs. Then, ~1×106 resuspended cells were subcutaneously injected into the nude mice (BALB/c female, 5 weeks old, n = 6). Mice were housed at ~20°C at a strictly controlled 12/12 light/dark cycle with free access to water and food. 28 days later, all mice were sacrificed. Solid tumors were then resected and weighed. Cells were covered with 25 nM FISH probes (Life Technologies, Shanghai, China) for 15 mins for hybridization according to the manufacturer’s guidelines and then dehydrated. The animal experiments were performed according to the Institutional Animal Care and Use Committee (IACUC) in Juxian Hospital of Traditional Chinese Medicine. The IACUC in Juxian Hospital of Traditional Chinese Medicine approved our experiments.
Statistics
Statistical analysis was performed using SPSS (version 16, SPSS, Inc., USA). Data were shown as mean ± SD. We used the Mann–Whitney test for comparison between two groups or ANOVA for comparison among multiple groups followed by Dunnett’s post hoc test. At least three replicates were included unless otherwise specified. P < 0.05 was considered statistically significant.
Results
Identification Of LINC00908 As A Potential HCC-Related lncRNA
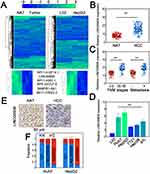
We performed lncRNA profiling to identify aberrantly expressed lncRNAs in HCC. The lncRNA profiling was first performed in normal adjacent tissues (NATs) and HCC tissues. To find potential oncogenic lncRNA in HCC, the lncRNAs which were upregulated in HCC samples were detected. Consequently, 71 significantly upregulated lncRNAs were identified (Figure 1A, left). The profiling was also conducted in L02/HepG2 cell lines and we found 91 significantly upregulated ones (Figure 1A, right). Overlapping study showed 6 novel lncRNAs related to HCC (Figure 1A, bottom and Table S2). Since LINC00908 displayed the highest fold increase (Table S2), we chose LINC00908 for further study. The LINC00908 genes was located on chromosome 18 (18q23) and the 5ʹ and 3ʹ rapid amplification of cDNA ends (RACE) and qRT-PCR data showed a dominant transcript and analysis of the transcript by Coding Potential Assessment Tool (CPAT) suggested minimal coding potential (Figure S1A–D). Northern blot showed that LINC00908 was demonstrably expressed in Huh7 and HepG2 HCC cell lines (Figure S1E). We noted that LINC00908 was substantially upregulated in HCC tissues compared with normal ones (Figure 1B). In addition, higher LINC00908 expression correlated with advanced TNM stages, metastasis and larger tumor size (Figure 1C and Table S3). Notably, LINC00908 did not show significant correlation with age and gender (Table S3). Consistently, LINC00908 expression was also elevated in multiple HCC cell lines (Figure 1D). In situ hybridization (ISH) data also showed higher LINC00908 expression in HCC tissues (Figure 1E). Fractionation results showed that LINC00908 revealed a dominant nuclear distribution (Figure 1F). These data suggested that LINC00908 might be associated with HCC with primary nuclear localization.
LINC00908 Advances HCC Progression In Vitro And In Vivo
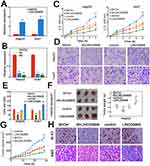
To further investigate the potentially oncogenic role of LINC00908, multiple in vitro experiments were performed. First, the overexpression and knockdown efficiency for LINC00908 were verified (Figure 2A and B). The results showed that LINC00908 overexpression or silence significantly altered LINC00908 levels in Huh7 and HepG2 cells (Figure 2A and B). Then, viability and migration assays were conducted. Overexpressing LINC00908 markedly promoted the viability of HepG2 and Huh7 cells while depleting LINC00908 attenuated viability (Figure 2C). The migratory capacity of Huh7 and HepG2 cells was also inhibited by LINC00908 knockdown and enhanced by higher LINC00908 abundance (Figure 2D and E). Furthermore, in vivo experiments were performed. The results suggested that LINC00908 overexpression substantially accelerated xenograft tumor growth, while LINC00908 knockdown significantly reduced tumor weight (Figure 2F). Consistently, lentiviral LINC00908 overexpression significantly increased tumor volumes, whereas silencing LINC00908 suppressed tumor growth (Figure 2G). Meanwhile, the fraction of positive Ki-67 staining also increased with LINC00908 overexpression (Figure 2H). These results suggested that LINC00908 may promote the oncogenesis of HCC both in vitro and in vivo.
LINC00908 Interacts With Sox-4 In HCC Cells
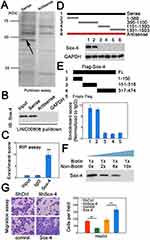
To elucidate the possible mechanisms of LINC00908-mediated oncogenic effect, we then explored the potential protein partner of LINC00908 in HepG2 cells using RNA pulldown. One specific band was identified (Figure 3A). This specifically enriched band was then subject to mass spectrometry, which showed 7 putative protein partners (Table S4). Immunoblots confirmed that LINC00908 could indeed interact with Sox-4 (Figure 3B). RNA immunoprecipitation (RIP) experiments showed that LINC00908 was enriched in Sox-4 pulldowns with antibody against Sox-4 (Figure 3C). A series of LINC00908 mutants were constructed to identify the binding regions to Sox-4. We found that the 1–389 nt fragment could physically interact with Sox-4 (Figure 3D). Furthermore, RIP assay demonstrated that N-terminal domain of Sox-4 was responsible for LINC00908 binding (Figure 3E). Competitive binding assays further confirmed the interaction between LINC00908 and Sox-4 (Figure 3F, biotin-labeled and unlabeled Sox-4 was used). As a result, silencing Sox-4 attenuated the migration of HepG2 cells, whereas Sox-4 overexpression enhanced the migration (Figure 3G, the efficiencies of Sox-4 silence or overexpression was verified in Figures S2A and B). Consistently, depleting Sox-4 decreased the viability of HCC cells, whereas Sox-4 overexpression dramatically enhanced the viability (Figure S3). These results demonstrated that LINC00908 could physically interact with Sox-4 to exert its oncogenic function.
LINC00908-Sox-4 Interaction Inhibits Proteasomal Degradation Of Sox-4
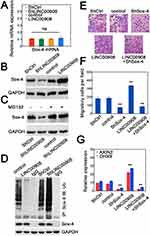
We then investigated the potential mechanism of LINC00908-mediated oncogenic effect by interaction with Sox-4. We measured transcript expression of Sox-4 and found that changing LINC00908 levels did not affect Sox-4 mRNA levels (Figure 4A). However, LINC00908 overexpression could elevate the abundance of Sox-4 at protein levels (Figure 4B). Silencing LINC00908 consistently decreased Sox-4 protein expression (Figure 4B). Moreover, the effect of reduction in Sox-4 expression with LINC00908 knockdown was strongly attenuated when proteasome inhibitor MG132 was added (Figure 4C), suggesting that LINC00908 could repress proteasomal degradation of Sox-4. We next investigated whether LINC00908 could influence Sox-4 ubiquitination. As expected, LINC00908 overexpression markedly decreased the ubiquitin ligation of Sox-4, whereas LINC00908 silence strongly increased Sox-4 ubiquitination (Figure 4D). Increasing LINC00908 expression promoted the migration of HepG2 cells, while silencing Sox-4 diminished the migratory capacity (Figure 4E and F). Notably, simultaneous LINC00908 overexpression and Sox-4 knockdown also substantially inhibited the migration to a similar extent when only Sox-4 was silenced (Figure 4E and F). The expression of two Sox-4 downstream genes (AXIN2 and DHX9) was also upregulated with LINC00908 overexpression and reduced when LINC00908 was silenced (Figure 4G). Similarly, Sox-4 knockdown strongly counteracted the effect of LINC00908 overexpression (Figure 4G). These results demonstrated that the interaction between LINC00908 and Sox-4 could protect Sox-4 from proteasomal degradation to increase the stability of Sox-4.
Discussion
In this study, we verified that a novel intergenic lncRNA termed LINC00908 could promote HCC viability and migration. Furthermore, LINC00908 also facilitated HCC xenograft tumor growth. Lowering LINC00908 expression could inhibit malignant phenotypes from in vivo and in vitro data. The oncogenic effect of LINC00908 was largely ascribed to its association with Sox-4. The LINC00908-Sox-4 interaction markedly increased Sox-4 stability through reducing ubiquitin-mediated proteasomal degradation. Therefore, all data supported an oncogenic role of LINC00908 in HCC.
lncRNAs are critically involved in numerous biological processes including oncogenesis.14,15 lncRNAs can fulfill its role via interaction with RNA, DNA or protein.7,8 For example, the loss of TSLNC8 is significantly enriched in HCC and TSLNC8 physically interacts with transketolase (TKT)/signal transducer and activator of transcription 3 (STAT3) to regulate the phosphorylation status of STAT3.16 lncRNA lncSox4 can interact with Sox-4 promoter through high-sequence complement to recruit STAT3 to Sox-4 promoter and induce Sox-4 expression.9 The lncRNA TNRC6C-AS1 can form a complex with miR-129-5p/UNC-5 netrin Receptor B (UNC5B) and serves as a competing endogenous RNA to promote thyroid cancer progression.17 Meanwhile, TNRC6C-AS1 also functions as a long noncoding antisense RNA by targeting TNRC6C to facilitate the aggressiveness of papillary thyroid cancer.18 In the current work, through RNA pulldown and mass spectrometry, we have identified that LINC00908 could physically interact with Sox-4 and dramatically attenuate the degradation of Sox-4. Therefore, we argued that the oncogenic role of LINC00908 might be ascribed to Sox-4 stabilization.
Sox-4 is a well-known transcription factor and actively involved in differentiation and proliferation.19 Sox-4 represses p53-mediated apoptosis in HCC by interaction with p53 via the high-mobility group (HMG) box domain to inhibit p53-mediated transcription.20 In the current work, we found that LINC00908 physically associated with the Sox-4 construct covering 1–150 residuals which also encompasses the HMG box domain. Therefore, Sox-4 might exert its oncogenic role through multiple strategies. Notably, the liver tumor-initiating cells (TICs) greatly contribute to the occurrence and malignancy of HCC.21 A recent report has demonstrated that lncRNA LncSox4 can recruit STAT3 to the promoter region of Sox-4 and activate Sox-4 transcription.9 Sox-4 protein is undoubtedly required for TIC stemness and highly expressed Sox-4 protein guarantees TIC self-renewal in liver TICs.9,21 Therefore, elevated expression of Sox-4 positively correlates with the progression and clinical severity in HCC.9 Consistently, a tumor-suppressive microRNA miR-449 can suppress HCC development by targeting Sox-4.22 Furthermore, miR-129-2 hypermethylation in HCC tumor tissues enhances Sox-4 expression and β-catenin/trans-acting T cell factors (TCF) activity.23 All these data have supported an oncogenic role of Sox-4 during HCC progression. Our study showed that LINC00908 could stabilize Sox-4 by protecting Sox-4 from proteasomal degradation in HCC and therefore establish a novel link between lncRNAs and HCC oncogenesis.
Notably, we do not exclude the possibility that LINC00908 could serve as a competing endogenous RNA to promote HCC development. Furthermore, whether LINC00908 could form complex with specific promoter regions through complementary sequences remains to be determined.
Conclusion
Collectively, we have found that a novel intergenic lncRNA LINC00908 is highly expressed in HCC and contributes to HCC progression. LINC00908 interacts with Sox-4, reduces ubiquitination of Sox-4 and induces Sox-4 stabilization. Therefore, LINC00908-Sox-4 axis might not only provide a novel marker for HCC but also serve as a putative target to eradicate liver cancer.
Author contributions
All authors contributed to conception and design, acquisition of data, interpretation of data, drafting or revising the article, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Funding
This work is supported by Innovation Program of Shandong Province (No. BS2017SF024).
Disclosure
The authors declare no competing financial interests.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10(2):99–111. doi:10.1016/j.ccr.2006.06.016
3. Rapisarda V, Loreto C, Malaguarnera M, et al. Hepatocellular carcinoma and the risk of occupational exposure. World J Hepatol. 2016;8(13):573–590. doi:10.4254/wjh.v8.i13.573
4. Goh GB, Chang PE, Tan CK. Changing epidemiology of hepatocellular carcinoma in Asia. Best Pract Res Clin Gastroenterol. 2015;29(6):919–928. doi:10.1016/j.bpg.2015.09.007
5. Necsulea A, Soumillon M, Warnefors M, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–640. doi:10.1038/nature12943
6. Jain S, Thakkar N, Chhatai J, Bhadra MP, Bhadra U. Long non-coding RNA: functional agent for disease traits. RNA Biol. 2016;14(5):1–14. doi: 10.1080/15476286.2016.1172756
7. Atianand MK, Caffrey DR, Fitzgerald KA. Immunobiology of long noncoding RNAs. Annu Rev Immunol. 2017;35:177–198. doi:10.1146/annurev-immunol-041015-055459
8. Guttman M, Rinn JL. Modular regulatory principles of large non-coding RNAs. Nature. 2012;482(7385):339–346. doi:10.1038/nature10887
9. Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598. doi:10.1038/ncomms12598
10. Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37(30):4094–4109. doi:10.1038/s41388-018-0250-z
11. Ding CH, Yin C, Chen SJ, et al. The HNF1alpha-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol Cancer. 2018;17(1):63. doi:10.1186/s12943-018-0813-1
12. Zhan M, He K, Xiao J, et al. LncRNA HOXA11-AS promotes hepatocellular carcinoma progression by repressing miR-214-3p. J Cell Mol Med. 2018;22:3758–3767. doi:10.1111/jcmm.2018.22.issue-8
13. Song L, Zhang S, Duan C, et al. Genome-wide identification of lncRNAs as novel prognosis biomarkers of glioma. J Cell Biochem. 2019. doi:10.1002/jcb.29259
14. Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi:10.1038/nrg3606
15. Wang F, Xie C, Zhao W, Deng Z, Yang H, Fang Q. Long non-coding RNA CARLo-5 expression is associated with disease progression and predicts outcome in hepatocellular carcinoma patients. Clin Exp Med. 2017;17(1):33–43. doi:10.1007/s10238-015-0395-9
16. Zhang J, Li Z, Liu L, et al. Long noncoding RNA TSLNC8 is a tumor suppressor that inactivates the interleukin-6/STAT3 signaling pathway. Hepatology. 2018;67(1):171–187. doi:10.1002/hep.29405
17. Hou S, Lin Q, Guan F, Lin C. LncRNA TNRC6C-AS1 regulates UNC5B in thyroid cancer to influence cell proliferation, migration, and invasion as a competing endogenous RNA of miR-129-5p. J Cell Biochem. 2018;119:8304–8316. doi:10.1002/jcb.26868
18. Muhanhali D, Zhai T, Jiang J, Ai Z, Zhu W, Ling Y. Long non-coding antisense RNA TNRC6C-AS1 is activated in papillary thyroid cancer and promotes cancer progression by suppressing TNRC6C expression. Front Endocrinol (Lausanne). 2018;9:360. doi:10.3389/fendo.2018.00420
19. Lai YH, Cheng J, Cheng D, et al. SOX4 interacts with plakoglobin in a Wnt3a-dependent manner in prostate cancer cells. BMC Cell Biol. 2011;12:50. doi:10.1186/1471-2121-12-50
20. Hur W, Rhim H, Jung CK, et al. SOX4 overexpression regulates the p53-mediated apoptosis in hepatocellular carcinoma: clinical implication and functional analysis in vitro. Carcinogenesis. 2010;31(7):1298–1307. doi:10.1093/carcin/bgq072
21. Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275–291. doi:10.1016/j.stem.2014.02.006
22. Sandbothe M, Buurman R, Reich N, et al. The microRNA-449 family inhibits TGF-beta-mediated liver cancer cell migration by targeting SOX4. J Hepatol. 2017;66(5):1012–1021. doi:10.1016/j.jhep.2017.01.004
23. Chen X, Zhang L, Zhang T, et al. Methylation-mediated repression of microRNA 129-2 enhances oncogenic SOX4 expression in HCC. Liver Int. 2013;33(3):476–486. doi:10.1111/liv.12097
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.