Back to Journals » OncoTargets and Therapy » Volume 12
The Long Non-Coding RNA SBF2-AS1 Exerts Oncogenic Functions In Gastric Cancer By Targeting The miR-302b-3p/E2F Transcription Factor 3 Axis
Authors Liang C, Yue C, Liang C, Ge H, Wei Z, Li G, Wu J, Huang H, Guo J
Received 31 March 2019
Accepted for publication 23 September 2019
Published 30 October 2019 Volume 2019:12 Pages 8879—8893
DOI https://doi.org/10.2147/OTT.S210697
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
This paper has been retracted
Chaojie Liang, 1,* Chaosen Yue, 2,* Chaowei Liang, 1 Hua Ge, 2 Zhigang Wei, 1 Guangming Li, 2 Jixiang Wu, 2 He Huang, 1 Jiansheng Guo 1
1Department of General Surgery, First Hospital/First Clinical Medical College of Shanxi Medical University, Taiyuan, Shanxi 030001, People’s Republic of China; 2Department of General Surgery, Beijing Tongren Hospital, Capital Medical University, Beijing 100730, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiansheng Guo; He Huang
Department of General Surgery, First Hospital/First Clinical Medical College of Shanxi Medical University, Taiyuan, Shanxi 030001 People’s Republic of China
Tel +86 150 3515 8467; +86 188 0126 1267
Email [email protected]; [email protected]
Background and aims: Studies show that the long non-coding RNA, SBF2-AS1, plays a critical role in cancer progression, but the role of SBF2-AS1 in gastric cancer has not been reported. Therefore, this study aimed to elucidate the mechanism of SBF2-AS1 in gastric cancer (GC).
Methods: A meta-analysis, based on the gene expression omnibus database and TCGA dataset was performed to explore the prognostic value of SBF2-AS1 in GC. RT-PCR was also conducted to investigate the clinicopathologic value of SBF2-AS1 in GC. The effect of SBF2-AS1 in GC cell lines was conducted by gain or loss-of-function assays, and the SBF2-AS1 target gene was confirmed using a luciferase reporter assay and bioinformatics.
Results: SBF2-AS1 was overexpressed in GC tissues and cell lines, and SBF2-AS1 overexpression indicated poor overall survival and could serve as an independent prognostic factor. Moreover, knockdown of SBF2-AS1 inhibited cell growth, invasion, and metastasis, promoted apoptosis, and caused cell cycle arrest. Luciferase reporter and gain- or loss-of-function assays indicated that SBF2-AS1 acted as a competing endogenous (ceRNA) for microRNA (miR)-302b-3p, which blocked the inhibitory effect of miR-302b-3p on the E2F transcription factor 3 (E2F3).
Conclusion: SBF2-AS1 could be a potential diagnostic and prognostic biomarker in GC, and SBF2-AS1 accelerates tumor progression via the miR-302b-3p/E2F3 axis.
Keywords: SBF2-AS1, gastric cancer, miR-302b-3p, E2F3
Introduction
Gastric cancer (GC) is the third leading cause of cancer-related deaths worldwide and poses a significant threat to human health.1 According to GLOBOCAN, 780,000 cases of GC-related deaths occurred worldwide each year, for which GC ranked third in males and fifth in women.2 And, in China, approximately 290,000 people die of GC every year. The key to reducing the GC mortality lies in early diagnoses and comprehensive, effective treatments. Although the current advanced diagnostic techniques and comprehensive treatment strategies have greatly improved from earlier years, being able to diagnose patients with GC early continues to be challenging, and no significant improvements the prognoses of patients with advanced GC have occurred. Therefore, searching effective biomarkers and specific therapeutic targets is an urgent task for early diagnoses and effective treatments of GC patients.3
Long non-coding RNAs (long ncRNAs) are a class of single-stranded RNA molecules over 200 nucleotides in length that do not encode proteins. At first, researchers considered that lncRNAs were a by-product of the transcription process,4 but in recent years, additional evidence suggests that lncRNAs are dysregulated in a variety of tumors and are associated with tumor recurrences and poor prognoses.5,6 Usually, dysregulated lncRNA expression affects cellular biological functions, including cell proliferation, invasion, metastases, angiogenesis, and apoptosis resistance.7,8 And these lncRNAs are involved in tumor progression mainly through silencing tumor suppressor genes and activating oncogene expression.9,10
SBF2-AS1 has been shown to be involved in the development of tumors such as hepatocellular carcinoma,11 cervical cancer,12 esophageal squamous cell carcinoma,13 and non-small cell lung cancer (NSCLC).14 Lv et al14 reported that SBF2-AS1 was significantly upregulated in NSCLC and that SBF2-AS1 expression was related to lymph node metastasis and advanced TNM stages. Moreover, SBF2-AS1 downregulation inhibited cellular proliferation and metastasis of NSCLC cell lines, and Zhao et al15 reported similar results. Chen et al13 found that SBF2-AS1 was overexpressed in ESCC tissues, which upregulated SBF2-AS1 expression and was associated with tumor sizes and TNM stages, and that SBF2-AS1 promoted cellular proliferation and invasion by regulating CDKN1A expression. Li et al16 reported that lncRNA SBF2-AS1 acted as an oncogene via the micro RNA (miR)-140-5p/TGFBR1 axis in hepatocellular carcinoma, and a similar conclusion was reported in another study.11 So SBF2-AS1 plays a vital role in promoting tumor progression. However, the role of SBF2-AS1 in GC has not been elucidated; and therefore, this study aimed to explore the mechanism of SBF2-AS1 in GC through the gene expression omnibus (GEO) and the cancer genome atlas (TCGA) database, in addition to experiments in tumor tissues, and in vivo and in vitro validation studies.
Materials And Methods
Search Strategy And Study Selection
GEO profiles (http://www.ncbi.nlm.nih.gov/geoprofiles/) and datasets (http://www.ncbi.nlm.nih.gov/gds/) were searched to analyze SBF2-AS1expression in GC. We only collected on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array, HG-U133_Plus_2), which minimized impacts on heterogeneity and searched as of March 1, 2019. The key words for the searches were “SBF2-AS1” OR “Long noncoding RNA SBF2-AS1” OR “LncRNA SBF2-AS1” AND “gastric cancers” or “gastric neoplasm.”
Specimens And Cell Lines
GC and adjacent normal tissues were surgically collected from 93 patients at the Beijing Tongren Hospital, Capital Medical University (Beijing, China) between 2012 and 2016. No treatments were conducted before sample collection. Clinicopathologic information was collected including gender, age, TNM stage, lymph node and distant metastases, and tumor differentiation. This study was approved by the ethics committee of the Beijing Tongren Hospital and performed according to the Declaration of Helsinki. Written informed consent was obtained from all of the patients.
The human GC cell lines, AGS, MKN45, HGC27, and SGC7901 were purchased from the Cancer Hospital of the Chinese Academy of Medical Sciences (Beijing, China). The MKN28 and normal gastric (GES) cell lines were purchased from the Chinese Academy of Sciences (Shanghai, China). Cells were cultured with RPMI 1640 containing 10% fetal bovine serum (FBS) (Gibco, Grand Island, NY, USA), 100 U/mL penicillin, and 100mg/mL streptomycin (Invitrogen, Carlsbad, CA, USA) at 37 °C in 5% CO2.
Cell Transfection
The SBF2-AS1 short-hairpin loop RNAs (shRNAs), SBF2-AS1 wild-type plasmids, SBF2-AS1 mutant plasmids, pCMV-E2F3 plasmids, and empty vectors were constructed by BIOREE Technology (Beijing, China). The micro RNA (miRNA) mimics and inhibitors were synthesized by BIOREE Technology (Beijing, China). The Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used for cell transfections according to the manufacturer’s instructions. SBF2-AS1 sh1 sense (204 site), 5ʹ-CAGAAGGAGUCUACUGCUAAG-3ʹ, and antisense, 5ʹ-UAGCAGUAGACUCCUUCUGG G-3ʹ, SBF2-AS1 sh2 sense (1021 site), 5ʹ-GCAAGCCUGCAUGGUACAUTT-3ʹ, and antisense, 5ʹ-AUGUACCAUGCAGGCUUGCTT-3ʹ. miR-302b-3p mimics (UAAGUGCUUCCAUGUUUUAGUAG) and normal control (NC) mimics (UUCUCCGAACGUGUCACGUTT)
RNA Extraction And qRT-PCR Analysis
The TRIzol reagent (Invitrogen, Karlsruhe, Germany) was used to extract the total RNA from the collected samples following the manufacturer’s instruction. The expression levels were confirmed using the Fast Start Universal SYBR Green Master (Roche, USA). We used Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 to normalize expression. 2−ΔΔCt was used for data analysis. The primers are shown in Table 1.
 |
Table 1 The Primers Of lncRNA And mRNAs |
Cell Viability Assay
Cell viability was detected using a CCK-8 assay according to the manufacturer’s instructions (Dojindo Laboratories, Kumamoto, Japan). Cells were seeded in 96-well plates (1×103/well). A total of 10 μL of the CCK-8 solution were added into the cultures, and then the cultures were incubated for 2 hrs at 37°C. We then measured the spectrophotometric absorbance at 450 nm. All experiments were performed in triplicate.
Xenograft Experiments
Animal studies were based on the Guide for the Care and Use of Laboratory Animals and conducted in accordance with the protocol approved by the Shanxi Medical University Application for Laboratory Animal Welfare and Ethical Committee. Six-week-old BALB/c nude mice were used for this study, and MKN28 cells (1×107 cells/mouse) from different groups were injected subcutaneously into the left flank of the mice. After three weeks, the mice were sacrificed, and tumor weights and volumes were evaluated.
The Wound Healing Assay
The wound healing assay was used to investigate cell migration. For this assay, cells were cultured at a density of 1 × 105/mL in serum-free DMEM media and 6-well plates. A wound was made along the center of each well, and the cell debris gently washed off with PBS. A microscope with an attached camera was used to acquire the photomicrographic images at 0 and 24 hrs. All experiments were performed in triplicate.
The Transwell Assay
The effect of SBF2-AS1 on cell invasion was investigated using the transwell chamber system (8 μm pore size; Costar) with Matrigel (BD Biosciences, San Jose, CA, USA). Briefly, 1×105 cells in serum-free medium were added into the upper chamber of the transwell with Matrigel. Six hundred microliters media supplemented with 10% FBS was added to the lower chamber. After 24 hrs, the non-invading cells were removed, and the cells that migrated to the bottom of the membrane were stained with crystal violet. Then, the cells were counted in 5 randomly selected fields under the microscope at 200× magnification.
Apoptosis And The Cell Cycle Analysis
Fluorescein isothiocyanate (FITC)-Annexin V staining was used for the apoptosis analysis. After double-staining with Annexin V-FITC and propidium iodide (PI) according to the manufacturer, the cells were analyzed with a flow cytometer (BD Accuri C6 flow cytometer, BD Biosciences). The percentage of Annexin V- and PI-positive cells were analyzed using CellQuest software (BD Biosciences). Cells incubated with RNase and stained with PI were used for cell cycle analysis according to the flow cytometry protocol.
Luciferase Reporter Analysis
pGL3-luc-SBF2-AS1 was constructed by inserting the wild-type (WT) or mutant (Mut) of the SBF2-AS1 or E2F3 3ʹ-UTR sequence, which contains the miR-302b-3p targeting sites into the pGL3 promoter vector (BIOREE, Beijing, China). For reporter assays, MKN28 and HGC27 cells (3×104/well) were seeded into 24-well plates and cultured overnight. The miRRiboTM mimics or miR-RiboTM negative control and the Mut reporter plasmid were used to co-transfect the cells using Lipofectamine 2000 (Invitrogen). The dual-luciferase reporter assay system (Promega) was used to analyze the relative luciferase activity, and Renilla luciferase was used as the internal control.
Western Blotting
The antibodies, E2F3 (1:1,000, Abcam, Cambridge, UK), N-cadherin (1:1,000, CST), vimentin (1:1,000, Cell Signaling Technology, Danvers, MA, USA), E-cadherin (1:1,000, Cell Signaling Technology, Danvers, MA, USA), and GAPDH (1:1,000, Cell Signaling Technology, Danvers, MA, USA) were used for Western blot analysis. Total protein was extracted from GC cells using the radioimmunoprecipitation assay (RIPA) buffer (Thermo Fisher Scientific) and bicinchoninic acid (BCA) Kit (Cwbiotech, Beijing, China) to quantify protein concentrations. Ten percent sodium dodecyl sulfate-polyacrylamide gel electrophoresis was used to separate proteins, which were transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). Five percent non-fat milk in Tris-buffered saline with Tween-20 (TBST) was added to the membrane for 1 hr to block non-specific protein interactions. Then, the appropriate individual primary antibody was used to incubate with the membranes overnight at 4°C. The membranes were washed three times with 1 × TBST, and primary antibody binding of the membranes was detected by incubating with appropriate secondary antibodies at room temperature for 50 mins. Eventually, an enhanced chemiluminescence system (EMD Millipore) was used to detect signal expression.
Statistical Analysis
The GraphPad Prism 6 software (GraphPad Software Inc., La Jolla, CA, USA), SPSS 22.0 software (IBM, Armonk, NY, USA) and STATA 14.2 (StataCorp LLC., College Station, TX, USA) were used for the statistical analyses. All experiments were performed in triplicate, and the data are represented as the mean ± SD. Comparisons among all groups were analyzed using the one-way analysis of variance test or unpaired Student’s t-test.
Results
A Meta-Analysis Examining The Expression And Prognostic Value Of lncRNA SBF2-AS1
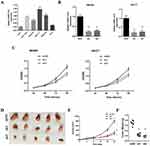
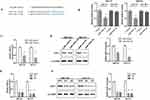
We first explored SBF2-AS1 expression levels in GC tissues using a meta-analysis, as shown in Table 2. Five datasets with 545 cancer tissues and 106 non-cancer tissues were enrolled from the GEO database and TCGA dataset. The pooled results indicated that SBF2-AS1 expression was overexpressed in cancer tissues (standardized mean difference [SMD] = 5.65, 95% CI = 4.16–7.15, P < 0.001, Figure 1A), and next we analyzed the prognostic value of SBF2-AS1, as shown in Table 3. Five studies with 987 patients were included in this meta-analysis, and the pooled hazard ratios (HRs) showed that high SBF2-AS1 expression was related to poor overall survival (OS) (HR = 1.48, 95% CI = 1.19–1.78, P < 0.01, Figure 1B). Together, the meta-analysis indicated that SBF2-AS1 was overexpressed in cancer tissues and that SBF2-AS1 overexpression was correlated with poor OS. Next, we performed a receiver operator characteristic (ROC) curve to evaluate the diagnostic value of SBF2-AS1 expression from the TCGA dataset (Figure 1C). The area under the ROC curve (AUC) was 0.72 (95% CI, 0.63–0.80), indicating that SBF2-AS1 could be a reliable biomarker for the diagnosis and prognosis of gastric cancer.
 |
Table 2 Characteristics Of Studies Based On Affymetrix Human Genome U133 Plus 2.0 |
 |
Table 3 OS Of Characteristics Of Studies Based On Affymetrix Human Genome U133 Plus 2.0 |
lncRNA SBF2-AS1 Is Overexpressed In GC Tissues And Associated With Poor Prognoses
Next, we explored SBF2-AS1 expression using qRT-PCR. SBF2-AS1 expression was higher in tumor tissues compared with the adjacent non-cancer tissues (Figure 1D). Then, we used the median SBF2-AS1 expression to divide the expression into high and low groups. To begin, we analyzed the relationship between SBF2-AS1 expression and clinicopathologic features, as shown in Table 4. SBF2-AS1 expression was related to tumor invasion depth and TNM stage, but not to gender, age, differentiation, or lymph node and distant metastases. A Kaplan-Meier analysis was then performed, and the log rank test demonstrated that upregulated SBF2-AS1 expression was related to shorter OS (Figure 1E, P < 0.001). Moreover, we evaluated the prognostic value of SBF2-AS1 using multivariate Cox analyses (Table 5). The results showed that SBF2-AS1 could be used as an independent prognostic factor to determine OS (HR=4.45, 95% CI: 2.013–9.837).
 |
Table 4 Expression Of SBF2-AS1 And Clinicopathological Parameters In GC |
 |
Table 5 Univariate And Multivariate Analysis Of Factors Associated With Overall Suvival |
lncRNA SBF2-AS1 Is Overexpressed In GC Cell Lines, And SBF2-AS1 Knockdown Suppresses The Proliferative Capacity Of GC Cell in vivo and in vitro
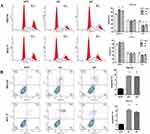
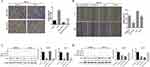
To investigate SBF2-AS1 function in GC, we detected SBF2-AS1 expression in GC cell lines and found that SBF2-AS1 was upregulated in GC cell lines compared with the normal GES cell line (Figure 2A), and HGC27 and MKN28 had the highest expression, so we chose the HGC27 and MKN28 for further research. We transfected the HGC27 and MKN28 cells with sh-SBF2-AS1, and as shown in Figure 2B, SBF2-AS1 expression was significantly downregulated. The CCK-8 assay (Figure 2C) was conducted to evaluate GC cell proliferation, and the results indicated that SBF2-AS1 downregulation suppressed the proliferative capability of GC cells,and we also performed BrdU assay to dectect the proliferation of GC cells, the results were similar to CCK-8 assay (Figure S1A). Next, we explored the role of SBF2-AS1 in vivo by injecting HGC27-sh1, HGC27-sh2, and HGC27-sh-NC cells into nude mice (Figure 2D), and the results showed that tumor volumes were significantly decreased in the sh1 and sh2 groups compared with the shNC group (Figure 2E) and that tumor weights were similar to tumor volumes (Figure 2F). We also wanted to determine the effect of SBF2-AS1 on the cell cycle and apoptosis (Figure 3A and B). The results indicated that SBF2-AS1 downregulation could promote apoptosis and arrest GC cell cycle arrest in the G0/G1 phase. Thus, the in vivo and in vitro validation demonstrated that proliferation was inhibited by downregulating SBF2-AS1 expression in gastric cancer cell lines.
SBF2-AS1 Knockdown Suppressed The Migration And Invasion Of GC Cells And The Epithelial-Mesenchymal Transition (EMT)
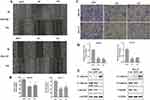
To explore the effect of SBF2-AS1 on migration and invasion in GC cell lines, wound healing and transwell assays were conducted using HGC27 and MKN28 cell lines. As shown in Figure 4A and B, SBF2-AS1 downregulation reduced cell migration in the wound healing assay, similar to what was seen in the transwell assay (Figure 4C and D), where cell invasion decreased in the sh groups compared with the normal groups, which indicated that SBF2-AS1 promotes the migration and invasion of GC cells. In addition, we conducted Western blot analysis to study the effects of SBF2-AS1 on the EMT, which included E-cadherin, N-cadherin, and Vimentin. The results (Figure 4E) showed that the epithelial-related protein, E-cadherin had increased expression when SBF2-AS1 was downregulated, and the mesenchymal-related protein was decreased. Therefore, SBF2-AS1 promoted the EMT process.
SBF2-AS1 Is A Target Of miR-302b-3p
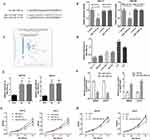
Accumulating evidence demonstrated that lncRNAs could be competing endogenous RNAs (ceRNA) for miRNAs,17,18 so we explored whether SBF2-AS1 could sponge miRNAs to promote GC progression. The bioinformatics tool, Starbase v3.0 software, was used to predict the potential ceRNA interaction with SBF2-AS1. For this experiment, miR-302b-3p was identified; a potential binding site that is shown in Figure 5A. We performed a luciferase assay (Figure 5B), which showed that miR-302b-3p overexpression significantly inhibited luciferase activity of the wild type SBF2-AS1(SBF2-AS1-WT), but not the mutant SBF2-AS1(SBF2-AS1-MUT). Bioinformatic analyses (Figure 5C) showed that SBF-AS1expression was negatively correlated with miR-302b-3p expression in the TCGA dataset. Besides, we also detected miR-302b-3p expression in GC cell lines, and miR-302b-3p expression was downregulated in GC cell lines compared with the normal gastric cell lines (Figure 5D). Moreover, as shown in Figure 5E, SBF2-AS1 downregulation increased miR-302b-3p expression, while miR-302b-3p downregulation upregulated SBF-AS1 expression, and miR-302b-3p mimics downregulated SBF2-AS1 expression (Figure 5F). Next, we performed the CCK-8 assay, and the results (Figure 5G) indicated that miR-302b-3p overexpression significantly suppressed the proliferative capability of the GC cells, and miR-302b-3p inhibitors promoted the proliferative capacity of the MKN28 and HGC27 cells. In addition, miR-302b-3p downregulation reversed the suppressed cell proliferation that was caused by silencing SBF2-AS1 expression (Figure 5H).
The E2F Transcription Factor 3 Is A Target Gene Of miR-302b-3p In GC Cells
To explore the potential target gene of miR-302b-3p, starbase v3.0 software (http://starbase.sysu.edu.cn/index.php) was used, and we identified E2F3 as the target for further research. The potential binding site between miR-302b-3p and E2F3 3ʹUTR is shown in Figure 6A. The luciferase reporter assay was performed to confirm this prediction, and the results (Figure 6B) showed that miR-302b-3p overexpression could significantly suppress the luciferase activity of the E2F3 3ʹ-UTR wild type but not E2F3 3ʹ-UTR mutant. Moreover, E2F3 mRNA and protein expression were remarkably reduced after miR-302b-3p was overexpressed in GC cell lines (Figure 6C and D). And, E2F3 mRNA and protein expression were significantly decreased after silencing SBF2-AS1 (Figure 6E and F).
Next, we used functional trials to verify this hypothesis with the transwell assay (Figure 7A), which showed that cell migration was enhanced by E2F3 overexpression, miR-302b-3p-mimics were able to suppress HGC27 cell migration, and miR-302b-3p inhibited the E2F3 effects on cell migration. Similarly, the E2F3 effects on invasion (Figure 7B) were also inhibited by miR-302b-3p. And, SBF2-AS1 overexpression using SBF2-AS1 wild-type plasmids could rescue E2F3 expression levels in stable SBF2-AS1-sh1, MKN28, and HGC27 cells; however, SBF2-AS1 mutant plasmids did not have that effect (Figure 7C), and miR-302b-3p knockdown reversed the reduced E2F3 expression (Figure 7D). Moreover, we also detected the expression of N-cadherin, E-cadherin and vemintin when overexpressed miR-302b-3p and E2F3, the results (Figure S1B) showed that the mesenchymal-related protein was decreased when overexpression of miR-302b-3p, and overexpression of E2F3 could rescue the expression of the mesenchymal-related protein such as Vimentin and N-cadherin. Thus, demonstrating that SBF2-AS1 functioned through the miR-302b-3p/E2F3 axis.
Discussion
Recently, it was shown that lncRNAs play an important role in the development of gastric cancer.19,20 In this study, we explored the role of SBF2-AS1 in GC. First, we performed a meta-analysis of the GEO database to find that SBF2-AS1 is highly expressed in gastric cancer and found that high SBF2-AS1 expression was associated with poor prognoses in gastric cancer patients. Subsequently, we found that SBF2-AS1 was overexpressed in gastric cancer tissues using RT-PCR and that SBF2-AS1 was found to be a reliable biomarker through ROC analyses. High SBF2-AS1 expression was associated with lymph node metastases, the depth of tumor invasion, and the TNM stage. Univariate analysis found that SBF2-AS1 was associated with a poor prognosis. Multivariate analysis found that SBF2-AS1 could be used as an independent factor for gastric cancer prognoses. Thus, we concluded that SBF2-AS1 could be used as a reliable molecular biomarker to determine prognoses in GC patients.
Next, we explored the biologic functions of SBF2-AS1 long ncRNAs in gastric cancer. RT-PCR results indicated that SBF2-AS1 was highly expressed in gastric cancer cell lines. Using HGC27 and MKN28 cells for further studies, we found that silencing SBF2-AS1 significantly inhibited GC cellular proliferation, invasion, and metastatic ability, promoted apoptosis, and arrested GC cells in the G0/G1 phase of the cell cycle. Tumorigenic abilities were also inhibited by silencing SBF2-AS1 expression, in vivo. An increasing number of studies found that lncRNAs play a role in the inhibition of tumorigenic abilities by competitively sponging miRNAs,21,22 and several studies have indicated that SBF2-AS1 can function as a tumor promoter by acting as a ceRNA. For example, Li et al16 indicated that SBF2-AS1 promotes hepatocellular cancer progression by competitively sponging miR-140-5p to regulate TGFBR1. To further elucidate the molecular mechanisms of SBF2-AS1 in gastric cancer, we performed a bioinformatics analysis, which predicted that SBF2-AS1 targets miR-302b-3p for binding, while TCGA database analyses found that SBF2-AS1 was negatively correlated with miR-302b-3p expression in GC. Moreover, the luciferase assays confirmed that miR −302b-3p mimics could significantly reduce luciferase activity of SBF2-AS1 WT but not SBF2-AS1 MUT. Silencing SBF2-AS1 increased miR-302b-3p expression. SBF2-AS1 expression was also affected by overexpression or knockdown of miR-302b-3p, miR-302b-3p expression, which significantly reversed the ability of SBF2-AS1 to inhibit tumor cell proliferation in GC cell lines. Therefore, these studies showed that SBF2-AS1 could act as a ceRNA by competitively sponging miR-302b-3p in gastric cancer cells.
Then, we explored how miR-302b-3p functions in gastric cancer. Previous studies23 found that miR-302b-3p is expressed at low levels in gastric cancer and can exert anti-cancer effects by targeting IGF-1R. Similar to previous studies, we also detected miR-302b-3p expression in GC cell lines using RT-PCR and found that miR-302b-3p expression was lower in GC cells than in normal gastric endothelial cells. Bioinformatics analysis suggested that E2F3 could be one of the target genes of miR-302b-3p in gastric cancer. Researchers24–27 have demonstrated that E2F3 is overexpressed in gastric cancer and can promote the development of gastric cancer. We found that miR-302b-3p mimics could significantly reduce the luciferase activity of WT E2F3 3ʹ-UTR, but not of MUT E2F3 3ʹ-UTR. In addition, silencing or overexpressing miR-302b-3p could reverse E2F3 mRNA and protein expression; and furthermore, we performed gain-or-loss experiments to confirm that E2F3 is one of the target genes of miR-302b-3p.
The relationship between SBF2-AS1 and E2F3 was also analyzed. First, we found that E2F3 mRNA and protein expression was inhibited after silencing SBF2-AS1. Then, SBF2-AS1 overexpression in a cell line with stable and low SBF2-AS1 expression caused an increase in E2F3 expression, which could be attenuated with miR-302b-3p mimics. Together, these results indicated that SBF2-AS1 can exert a cancer-promoting effect by competitively sponging miR-302b-3p to target and regulate E2F3 expression.
In summary, our study found that SBF2-AS1 is overexpressed in gastric cancer tissues and cell lines and was associated with a poor prognosis. The mechanism of SBF2-AS1 in GC is shown in Figure 8. Silencing SBF2-AS1 can inhibit the proliferation, invasion, and metastasis of GC cells and can arrest GC cells in the G0/G1 phase of the cell cycle and promote apoptosis. In addition, we found that SBF2-AS1 could act as a ceRNA to target and regulate E2F3 expression by competitively sponging miR-302b-3p. This study provides the potential for a new therapeutic target that could be used as a diagnostic and prognostic marker of GC.
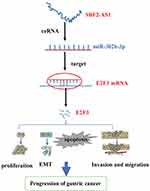 |
Figure 8 The diagram for the mechanism of SBF2-AS1 in the promotion of gastric cancer cell progression. |
Author Contributions
Jiansheng Guo and He Huang designed the study and performed the experiments; Chaojie Liang, Chaosen Yue, Hua Ge, Zhigang Wei, Guangming Li and Jixiang Wu performed the experiments and analyzed the data; Chaojie Liang and Chaosen Yue wrote the manuscript. All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi:10.3322/caac.21387
2. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi:10.3322/caac.21492
3. Hamashima C. Current issues and future perspectives of gastric cancer screening. World J Gastroenterol. 2014;20:13767–13774. doi:10.3748/wjg.v20.i38.13767
4. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi:10.1101/gad.1800909
5. Liang C, Zhang B, Ge H, et al. Long non-coding RNA CRNDE as a potential prognostic biomarker in solid tumors: a meta-analysis. Clin Chim Acta. 2018;481:99–107. doi:10.1016/j.cca.2018.02.039
6. Liang C, Qi Z, Ge H, et al. Long non-coding RNA PCAT-1 in human cancers: a meta-analysis. Clin Chim Acta. 2018;480:47–55. doi:10.1016/j.cca.2018.01.043
7. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi:10.1038/nrg2521
8. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21:1253–1261. doi:10.1038/nm.3981
9. Huang G, Wu X, Li S, et al. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci Rep. 2016;6:26524. doi:10.1038/srep26524
10. Chen Z, Yu C, Zhan L, et al. LncRNA CRNDE promotes hepatic carcinoma cell proliferation, migration and invasion by suppressing miR-384. Am J Cancer Res. 2016;6:2299–2309.
11. Zhang YT, Li BP, Zhang B, et al. LncRNA SBF2-AS1 promotes hepatocellular carcinoma metastasis by regulating EMT and predicts unfavorable prognosis. Eur Rev Med Pharmacol Sci. 2018;22:6333–6341. doi:10.26355/eurrev_201810_16044
12. Gao F, Feng J, Yao H, et al. LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol. 2019;47:776–782. doi:10.1080/21691401.2019.1577883
13. Chen R, Xia W, Wang X, et al. Upregulated long non-coding RNA SBF2-AS1 promotes proliferation in esophageal squamous cell carcinoma. Oncol Lett. 2018;15:5071–5080. doi:10.3892/ol.2018.7968
14. Lv J, Qiu M, Xia W, et al. High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res. 2016;35:75. doi:10.1186/s13046-016-0352-9
15. Zhao QS, Li L, Zhang L, et al. Over-expression of lncRNA SBF2-AS1 is associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci. 2016;20:3031–3034.
16. Li Y, Liu G, Li X, et al. Long non-coding RNA SBF2-AS1 promotes hepatocellular carcinoma progression through regulation of miR-140-5p-TGFBR1 pathway. Biochem Biophys Res Commun. 2018;503:2826–2832. doi:10.1016/j.bbrc.2018.08.047
17. Zheng J, Liu X, Wang P, et al. CRNDE promotes malignant progression of glioma by attenuating miR-384/PIWIL4/STAT3 axis. Mol Ther. 2016;24:1199–1215. doi:10.1038/mt.2016.71
18. Liu D, Zhu Y, Pang J, Weng X, Feng X, Guo Y. Knockdown of long non-coding RNA MALAT1 inhibits growth and motility of human hepatoma cells via modulation of miR-195. J Cell Biochem. 2017;119(2):1368–1380.
19. Zhang S, Xu J, Wang H, Guo H. Downregulation of long noncoding RNA LINC00460 expression suppresses tumor growth in vitro and in vivo in gastric cancer. Cancer Biomark. 2019;24:429–437. doi:10.3233/CBM-182177
20. Wang X, Kan J, Han J, et al. LncRNA SNHG16 functions as an oncogene by sponging MiR-135a and promotes JAK2/STAT3 signal pathway in gastric cancer. J Cancer. 2019;10:1013–1022. doi:10.7150/jca.29527
21. Xu L, Wei B, Hui H, et al. Positive feedback loop of lncRNA LINC01296/miR-598/Twist1 promotes non-small cell lung cancer tumorigenesis. J Cell Physiol. 2018;34(4):4563–4571.
22. Qin QH, Yin ZQ, Li Y, Wang BG, Zhang MF. Long intergenic noncoding RNA 01296 aggravates gastric cancer cells progress through miR-122/MMP-9. Biomed Pharmacother. 2018;97:450–457. doi:10.1016/j.biopha.2017.10.066
23. Guo B, Zhao Z, Wang Z, et al. MicroRNA-302b-3p suppresses cell proliferation through AKT pathway by targeting IGF-1R in human gastric cancer. Cell Physiol Biochem. 2017;42:1701–1711. doi:10.1159/000479419
24. Manicum T, Ni F, Ye Y, Fan X, Chen BC. Prognostic values of E2F mRNA expression in human gastric cancer. Biosci Rep. 2018;38. doi:10.1042/BSR20181264
25. Zhou X, Ji G, Ke X, et al. MiR-141 inhibits gastric cancer proliferation by interacting with long noncoding RNA MEG3 and down-regulating E2F3 expression. Dig Dis Sci. 2015;60:3271–3282. doi:10.1007/s10620-015-3782-x
26. Guo Y, Qi Y, Guo A, et al. miR-564 is downregulated in gastric carcinoma and targets E2F3. Oncol Lett. 2017;13:4155–4160. doi:10.3892/ol.2017.5964
27. Yang H, Wang L, Tang X, Bai W. miR-203a suppresses cell proliferation by targeting E2F transcription factor 3 in human gastric cancer. Oncol Lett. 2017;14:7687–7690. doi:10.3892/ol.2017.7199
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.