Back to Journals » OncoTargets and Therapy » Volume 13
The lncRNA LINC00691 Functions as a ceRNA for miRNA-1256 to Suppress Osteosarcoma by Regulating the Expression of ST5
Authors Wan D, Qu Y , Zhang L, Ai S, Cheng L
Received 27 June 2020
Accepted for publication 3 December 2020
Published 24 December 2020 Volume 2020:13 Pages 13171—13181
DOI https://doi.org/10.2147/OTT.S266435
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Takuya Aoki
Daqian Wan,1,2,* Yang Qu,3,* Lei Zhang,4,5 Songtao Ai,3 Liming Cheng1,2
1Department of Orthopedics, Tongji Hospital Affiliated to Tongji University School of Medicine, Shanghai, People’s Republic of China; 2Key Laboratory of Spine and Spinal Cord Injury Repair and Regeneration, Ministry of Education of the People’s Republic of China, Shanghai, People’s Republic of China; 3Department of Radiology, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, People’s Republic of China; 4Department of Orthopedics, The First Affiliated Hospital of Shandong First Medical University, Shandong, People’s Republic of China; 5Department of Orthopedics, Shandong Provincial Qianfoshan Hospital, Shandong University, Shandong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Songtao Ai
Department of Radiology, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, No. 639 Zhizaoju Road, Huangpu District, Shanghai, People’s Republic of China
Tel +86 18019790801
Email [email protected]
Liming Cheng
Tongji Hospital Affiliated to Tongji University School of Medicine, No. 389 Xincun Road, Putuo District, Shanghai, People’s Republic of China
Tel +86 13701959550
Email [email protected]
Introduction: Osteosarcoma is the most common primary malignant tumor in children and young patients. Although neoadjuvant chemotherapy and surgery could improve the prognosis of these patients, treatment outcomes are poor because of its low early diagnosis rate and high degree of malignancy as well as its tendency for early metastasis. In the field of osteosarcoma, lncRNAs have become a hot spot for studying the molecular mechanisms driving malignant biological characteristics and exploring effective treatment methods. An lncRNA is a long noncoding RNA lacking protein-encoding ability, and in its RNA form, it regulates various gene expression processes, such as epigenetic regulation, transcriptional regulation, and posttranscriptional regulation. LncRNAs play an important role in tumorigenesis and metastasis.
Methods: We used bioinformatics software to analyze the data in geo database. CCK-8 and Transwell were used to detect the effect of lncRNA LINC00691 on the proliferation and migration of osteosarcoma cells. The target gene of LINC00691 was detected by bioinformatics analysis and RNA pull down.
Results: In this study, we identified the lncRNA LINC00691 and confirmed its expression in osteosarcoma cells through GEO database analysis. Expression analysis showed that the levels of lncRNA LINC00691 in osteosarcoma cells were decreased compared to those of control cells. Overexpression of LINC00691 could inhibit the proliferation, migration, invasion, and induction of G1 cell cycle arrest in osteosarcoma cells, which was shown through in vitro and in vivo studies. Using bioinformatics analysis, RNA pull down experiments and luciferase reporter gene detection assays, we found that LINC00691 regulated ST5 expression by binding miR-1256. LINC00691 overexpression inhibited EMT by promoting the expression of E-cadherin and increasing the expression of ZEB1, Snail, and Fibronectin.
Conclusion: These results suggested that overexpressed LINC00691 promoted the expression of ST5 by regulating the function of miR-1256 through a ceRNA mechanism. The LINC00691/miR-1256/ST5 pathway plays an important role in the progression and metastasis of osteosarcoma and represents a good therapeutic target.
Keywords: osteosarcoma, ST5, lncRNA, miR-1256
Introduction
Osteosarcoma is one of the most common malignant bone tumors in children and adolescents under the age of 20; it accounts for approximately 5% of pediatric tumors.1–3 Osteosarcoma is one of the most common malignant tumors of bone and is derived from mesenchymal cells.4 The rapid growth of these tumors is due to the formation of tumor bone-like tissue and bone tissue, which occurs directly or indirectly through the cartilage stage. Osteosarcoma is still a high mortality disease in children and adolescents, but early detection and timely treatment have greatly improved the survival rate of patients with this disease. After the pathological diagnosis of osteosarcoma, early chemotherapy or radiotherapy is started. Resection of tumor tissue is an important step in the treatment of osteosarcoma. With the improvement of tumor surgical technology and the development of implant research, limb preservation therapy shows good therapeutic prospects. Consolidation chemotherapy or radiotherapy after tumor resection is very important for control of tumor metastasis and for improving the survival rate.2,5 Radical operation should be performed in the treatment of osteosarcoma. If conditions permit, local extensive resection and limb preservation can be performed. In addition, biopsies should be performed before amputation. Immunotherapy includes intravenous infusion of lymphocytes or interferon and transfer factors, but the curative effect of these treatments is still uncertain.6,7 Therefore, it is very important to study the molecular mechanism of osteosarcoma cell genesis and metastasis for the clinical diagnosis and treatment of osteosarcoma.
LncRNAs are functional RNA molecules with transcripts longer than 200 nucleotides.8,9 Early studies found that lncRNAs lack conserved open reading frames (ORFs), so they were considered “noise” with no biological characteristics. However, with further research, scholars have found that lncRNAs play key roles in epigenetic regulation, transcriptional regulation, posttranscriptional regulation, and nuclear transport, and they participate in cell histone modification, chromatin remodeling, DNA methylation, and gene activation or silencing. Most lncRNAs are transcribed by RNA polymerase II and then processed and modified into mature lncRNAs. A large number of lncRNAs exist in the nucleus, and a small number are in the cytoplasm. The main biological functions of lncRNAs are as follows: (1) as a signaling molecule, lncRNAs can generate tissue or cell specificity after they are activated; (2) lncRNAs can participate in chromosome silencing, genomic imprinting, chromatin modification, transcriptional activation, transcriptional interference, intranuclear transport, etc.; (3) lncRNAs can protect protein-coding genes in a variety of ways; (4) they contain highly efficient key sequences, which are similar to promoters and other regulatory factors, and they are more sensitive to structural and functional limitations than proteins; and (5) they can act in both cis and trans ways to bidirectionally regulate target genes.10–12
In recent years, it has been found that long noncoding RNAs are closely related to the biological processes of tumor development, invasion, and metastasis.13 H19 was the first lncRNA found to be associated with cancer. It is the imprinted gene product of insulin-like growth factor-2. Studies have found that H19 could promote the proliferation of cancer cells, inhibit apoptosis, promote angiogenesis, and increase cell tolerance of hypoxia.14,15 The abnormal expression of lncRNAs not only could participate in the process of tumor inhibition but also could participate in the process of carcinogenesis.16,17 It has been reported that the abnormal expression of lncRNAs is related to the occurrence of many kinds of tumors, such as breast cancer, colon cancer, and gastric cancer.18–20 In this study, we verified that a novel lncRNA, LINC00691, is associated with osteosarcoma through bioinformatics analysis and experimental detection.
In this study, we will examine the regulatory effect of lncRNA LINC00691 on osteosarcoma cells in vivo and in vitro and study the mechanism by which LINC00691 regulates osteosarcoma cells. This study will help us to improve our understanding of the function of lncRNAs in regulating osteosarcoma, help to develop new therapeutic strategies for osteosarcoma, and provide patients with improved treatments and quality of life.
Materials and Methods
Cell Culture
Osteosarcoma cells (U2OS, Saos-2, MG-63, and HOS) were cultured in DMEM medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 mg/mL streptomycin. Human osteoblast hFOB1.19 cells were cultured in DMEM/F-12 (1:1) medium containing 10% fetal bovine serum. Hff-1 (human foreskin fibroblast-1) and hBMSCs (human bone marrow stem cells) were cultured in α-MEM medium containing 10% fetal bovine serum, 100 U/mL penicillin, and 100 μg/mL streptomycin (from Carlsbad, GIBCO, California, USA). The cells were cultured in a humidified incubator at 37°C with 5% CO2. All cells were purchased from Cell bank of representative culture preservation Committee of Chinese Academy of Sciences.
Cell Transfection
Cells were transfected with Lipofectamine 3000 (Invitrogen; Thermo Fisher Scientific, Inc.) in accordance with the manufacturer’s instructions. A LINC00691 overexpression vector was constructed using a pLVX-IRES-Puro vector provided by SANGON Biotechnology Co., Ltd. An shRNA knockdown LINC00691 was purchased from Genechem Co., Ltd. (Shanghai, China). Mir-1256 mimics and inhibitors were purchased from Genepharma (Shanghai, China).
Real-Time Fluorescent Quantitative PCR Detection
Total RNA was extracted by TRIzol reagent, and RNA concentrations were detected by nanodrop. Total RNA (1 μg) was reverse transcribed with an RNA reverse transcription kit. Real-time quantitative PCR was performed using an SYBR Green RT qPCR Master Mix kit. Samples were prepared according to the instructions of the kit. The reaction conditions were as follows: program 1: 95°C, 30 s, 1 cycle; program 2: 95°C, 5 S, 50 cycles, 60°C, 34 s; program 3: 95°C, 5 s, 1 cycle, 65°C, 60 s, 97°C, 1 s; program 4: 42°C, 30 s, 1 cycle. The relative gene expression was calculated by the 2 - Δ CT method, and the mRNA expression was corrected based on the expression of GAPDH.
Cell Invasion Ability Detection
After DMEM was mixed with Matrigel 1:1, 50 μL of the mix was evenly spread in Transwell plates and placed in a 37°C incubator for 45 min. There were four groups of cells as follows: pLVX-Vector group, pLVX-LINC00691 group, pLKO.1-Vector group, and pLKO.1-LINC00691 group. The cells were inoculated into the Transwell upper chamber at a density of 20,000 cells per well. Serum-free medium was used in the upper chamber of the chamber, and 700 μL of medium containing 5% serum was added into the lower chamber. The cells were removed after 12 h of incubation in the incubator. The cells on the bottom of the Transwell were stained with crystal violet; then images were captured of the cells, and then they were counted.
CCK-8 Analysis
Osteosarcoma cells were inoculated into 96-well plates at a density of 400 cells per well. There were four groups of cells as follows: pLVX-Vector group, pLVX-LINC00691 group, pLKO.1-Vector group, and pLKO.1-LINC00691 group. Cell viability was detected by CCK-8 assay at 24, 48, 72, and 96 h after the cells were plated. Ten microliters of CCK-8 solution was added to each well, and then the cells were incubated in a 37°C incubator for 2 h. The absorbance at 450 nm was determined by an enzyme labeling method.
Colony Forming Assay
Osteosarcoma cells were inoculated into 6-well plates with 400 cells per well. There were four groups of cells: pLVX-vector group, pLVX-LINC00691 group, pLKO.1-vector group, and pLKO.1-LINC00691 group. The culture medium was changed every 3 days after inoculation. After 14 days of culture, the cells were fixed with 4% paraformaldehyde for 15 min. After fixation, 0.1% crystal violet staining solution was added to the dye for 5 min. Then, PBS was washed twice to remove nonspecific staining. After taking photos, the number of clones was determined.
Dual-Luciferase Reporter Experiment
Cells were seeded into 96-well plates at a density of 1000 cells per well. After 24 h of inoculation, lipofectamine was used. The luciferase reporter gene and miRNA were cotransfected into osteosarcoma cells in groups based on the following combinations: pmirGLO-LINC00691-wild-type (WT) and miRNA mimics NC; pmirGLO-LINC00691-wild-type (WT) and miR-1256 mimics; pmirGLO-LINC00691-mutant (MUT) and miRNA mimics; or pmirGLO-LINC00691-mutant (MUT) and miR-1256 mimics (Promega Corporation, Madison, WI, USA). After incubation at 37°C for 48 h, firefly and Renilla luciferase activity was determined using a dual-luciferase reporter analysis system (Promega Corporation, Madison, WI, USA) according to the manufacturer’s instructions. The firefly luciferase activity was normalized to that of Renilla.
RNA Pull Down
The cells were harvested, resuspended in nuclear separation buffer, placed on ice for 20 min, and mixed frequently. The nuclei were prepared by centrifugation at 2500 g for 15 min. RIPA buffer was used to resuspend the nucleus, and then it was divided into two parts (mock and pull down). The chromatin was mechanically sheared by homogenizing with a dounce homogenizer 15–20 times. After centrifugation, the nuclear membrane was separated from the fragments. Anti-MS2b antibody (20 μg) was added to the culture supernatant (10 mg) and was incubated for 2 h at 4°C overnight. Protein A/g beads (40 μL) were added to the mixture and incubated at 4°C for 1 h. The mixture was washed in RIPA buffer and then was washed with PBS. TRIzol was used to isolate the RNA.
Bioinformatics Prediction
The gene data for osteosarcoma cells were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) (GSE85537) for gene expression analysis. The target gene miR-1256 was identified using the TargetScan website (http://www.targetscan.org/vert_ 72/). The miRNAs that might bind to LINC00691 were identified using the miRcode website (http://www.mircode.org/).
Xenotransplantation of Tumor
All experimental procedures were based on the EU directive 2010/63/EU for animal experiments (http://ec.europa.eu/environment/chemicals/lab_animals/legislation_ en.htm). This experiment was approved by the ethics committee of Harbin Fifth Hospital (xhec-f-18-3). Six-week-old female BALB/C nude mice were purchased from Better Biotechnology Co., Ltd (Nanjing, China). Transfected cells (1 × 107) were suspended in 100 μL of PBS and then were inoculated subcutaneously into nude mice (5 mice in each group). After 3 weeks, all mice were sacrificed, and the tumors were removed and weighed.
Statistical Analysis
Student’s t-tests were used to calculate p values using SPSS 19.0. All data are expressed as the mean ± standard deviation (S.D.). P < 0.05 was considered to be statistically significant.
Result
The Expression of lncRNA LINC00691 is Decreased in Osteosarcoma Cells
Bioinformatics analysis using GEO database data information showed that the in situ expression of LINC00691 was decreased in lung metastasis compared with osteosarcoma (Figure 1A). The expression of LINC00691 in osteosarcoma cells (hBMSCs, HFF-1, and hFOB1.19 cells) was detected by qRT-PCR. The results showed that the expression of LINC00691 in osteosarcoma cells was decreased compared to its levels in hBMSCs, HFF-1, and hFOB1.19 cells (Figure 1B).
LINC00691 Regulates the Proliferation of Osteosarcoma Cells
According to the expression of LINC00691 in osteosarcoma cell lines, we selected U2OS and Saos-2 cells for subsequent functional and mechanistic analysis. LINC00691 expression in osteosarcoma cells was increased after transfection with pLVX-LINC00691 compared with the cells transfected with the pLVX-Vector (Figure 2A). In contrast, the expression of LINC00691 was decreased in osteosarcoma cells after transfection with pLKO.1-LINC00691 compared with cells in the pLKO.1-Vector group (Figure 2A). Cell viability was detected by CCK-8 assay, and the results showed that lncRNA LINC00691 overexpression suppressed the viability of osteosarcoma cells at 48 h, 72 h, and 96 h compared to the control group (Figure 2B). In contrast, lncRNA suppression promoted the viability of osteosarcoma cells at 48 h, 72 h, and 96 h (Figure 2B).
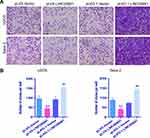
LINC00691 Regulates the Invasion of Osteosarcoma Cells
The effect of LINC00691 on the invasion of osteosarcoma cells was determined by a Transwell Matrigel test. The invasion ability of osteosarcoma cells in the pLVX-LINC00691 group was lower than that of the pLVX-Vector group (Figure 3A). However, the invasion ability of osteosarcoma cells in the pLKO.1-LINC00691 group was higher than that in the pLKO.1-Vector group (Figure 3B).
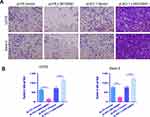
LINC00691 Regulates the Effect of Osteosarcoma Cells on the Invasion of HUVECs
Osteosarcoma cells could promote the invasion of HUVECs, so we assessed how the regulation of LINC00691 in osteosarcoma cells impacted the invasion of HUVECs using Transwell detection. The Transwell detection results showed that the invasion of HUVECs in the pLVX-LINC00691 group was lower than that of untreated HUVECs (Figure 4A and B). However, the invasion of HUVECs in the pLKO.1-LINC00691 group was higher than that of untreated HUVECs (Figure 4A and B).
LINC00691 Regulates EMT in Osteosarcoma Cells
EMT is closely related to tumor metastasis, so we detected if the expression of EMT-related genes in osteosarcoma cells was regulated by LINC00691. qRT-PCR detection results showed that LINC00691 overexpression promoted the expression of E-cadherin and inhibited the expression of ZEB1, Snail, and Fibronectin (Figure 5). In contrast, LINC00691 suppression inhibited the expression of E-cadherin and promoted the expression of ZEB1, Snail, and Fibronectin (Figure 5).
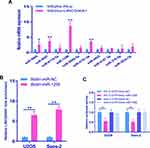
LncRNA LINC00691 Functions as a ceRNA of miR-1256 to Regulate Osteosarcoma Cells
At present, studies have shown that a ceRNA-related mechanism is one of the main mechanisms by which lncRNAs regulate cell function. Therefore, we used bioinformatics software to analyze miRNAs with the lncRNA LINC00691 binding site and then verified them by RNA pull down and dual-luciferase reporter analysis. MS2bs RNA pull down results showed that lncRNA LINC00691 could bind with miR-1256 (Figure 6A). Dual-luciferase reporter assays showed that lncRNA LINC00691 could bind with miR-1256 (Figure 6B). Additionally, we found in a biotin RNA pull down experiment that miR-1256 could bind with lncRNA LINC00691 (Figure 6C).
LncRNA LINC00691 Regulates the Expression of ST5 in a miR-1256-Dependent Manner
MiRNAs play a role in gene regulation mainly by binding with the target gene 3ʹ- UTR to inhibit the translation of the target gene. TargetScan showed the binding site of miR-1256 with the ST5 mRNA 3ʹ-UTR (Figure 7A). Dual-luciferase reporter assays showed that miR-1256 could bind with the 3ʹ-UTR of ST5 mRNA (Figure 7B). The expression of ST5 in osteosarcoma cells was detected by qRT-PCR. qRT-PCR results showed that there was less expression of ST5 in osteosarcoma cells than there was in hBMSCs (Figure 7C). There was no change in ST5 mRNA expression in osteosarcoma cells when miR-1256 was overexpressed or knocked down (Figure 7D). Next, a subcutaneous transplantation tumor model was established in nude mice. The results showed that the tumor volume of the pLVX-LINC00691 group was significantly smaller than that of the pLVX-vector group; in contrast, the tumor volume of the pLKO.1-LINC00691 group was significantly larger than that of the pLKO.1-vector group (Figure 7E).
Discussion
In recent years, the clinical treatment of osteosarcoma has encountered a bottleneck, and the effects of therapies have plateaued. With the rapid development and application of molecular biology technology, the molecular mechanisms of lncRNAs have been studied and clarified.21 LncRNAs not only can assist the early diagnosis and prognosis evaluation of osteosarcoma but also can regulate its biological characteristics and chemotherapy resistance, which means that lncRNAs are expected to become a new therapeutic target for osteosarcoma treatment. However, research on lncRNAs is also facing some challenges, such as the following: (1) the number of lncRNAs is huge, and the function and mechanism of lncRNAs are diverse; (2) it is difficult to predict the function of lncRNAs only by their sequence; and (3) at present, lncRNAs that have not been characterized may encode key proteins or polypeptides, which may affect the functional research of target lncRNAs. Despite these difficulties, lncRNA is still considered the “tomorrow star” in the field of cancer research. Exploring the impact of lncRNAs on osteosarcoma and their molecular mechanisms could be used in the screening of high-risk patients, reducing incidence rate and mortality, improving prognosis and survival rate, and providing new ideas and methods for clinical diagnosis and treatment. In this study, we found that LINC00691 was downregulated in osteosarcoma cell lines. Through overexpression and knockdown of LINC00691 in osteosarcoma cells, we found that LINC00691 can inhibit the proliferation, migration, and invasion of osteosarcoma cells and that LINC00691 can cause G1 phase arrest of osteosarcoma cells. LINC00691 can promote the proliferation of osteoma in vivo. We found that LINC00691 can regulate the function of miR-1256 and then mediate the expression of ST5. Our study is the first report on the regulatory effect of linc00691 on osteosarcoma and its regulatory mechanism.
In recent years, a variety of lncRNAs have been confirmed to be involved in the initiation and progression of osteosarcoma. LncRNAs sox2-ot, malat1, Thor, panda, and foxd2-as1 have been confirmed to be upregulated in tumor tissues, and they can regulate the proliferation, migration, and invasion of osteosarcoma cells; further, they may be used as biomarkers in the diagnosis of osteosarcoma.22–25 Bone marrow mesenchymal stem cells deliver lncRNA PVT1 into osteosarcoma cells by secreting exosomes, and lncRNA PVT1 promotes the expression of ERG by reducing the ubiquitination level of ERG, thus regulating the growth and metastasis of osteosarcoma cells.26 TUSC-7 suppresses the proliferation and migration of osteosarcoma cells and promotes the apoptosis of osteosarcoma cells by acting as a ceRNA of miR-211.27 In this study, we found that LINC00691 functions as a tumor suppressor gene that can inhibit the proliferation, migration, migration, and invasion of osteosarcoma cells.
A lncRNA is a linear RNA that does not encode a translated protein. A lncRNA could act as a sponge to buffer and inhibit the expression of its target genes by competing for intracellular miRNAs. It can play a role as a “miRNA sponge”. Some data show that miRNA sponges are broad regulators of miRNA activity in many eukaryotes.28,29 Previous studies have shown that lncRNAs can act as endogenous competitive RNAs (competing endogenous RNAs, ceRNAs); through competitive binding of common miRNAs, lncRNAs can regulate the expression of miRNA targets by downregulating miRNA expression and activity.30 LncRNAs have specific cell types, tissue types, developmental stages, and disease-specific expression patterns and locations. They may be effective natural miRNA sponges under some conditions.31 The lncRNA PCAT6 is an oncogene in osteosarcoma that functions as a ceRNA for miR-185-5p.32 The lncRNA HIF1A-AS2 promotes osteosarcoma cell proliferation and migration by acting as a ceRNA for miR-33b-5p to modulate SIRT6.33 The lncRNA MALAT1 is highly expressed in osteosarcoma tissues and cells, and it promotes cell growth and tumor progression by inhibiting miR-376a.34 The lncRNA MEG3 acts as an endogenous sponge by binding with miR-127, and ZEB1 is the target gene of miR-127. When ZEB1 is overexpressed, miR-127 is also overexpressed, which promotes cell growth and metastasis; additionally, miR-127 activates the JNK and Wnt signaling pathways. In conclusion, their research results showed that the lncRNA MEG3 promoted the growth and metastasis of osteosarcoma cells in vitro through miR-127.35 In this study, through bioinformatics analysis, we found that LINC00691 and multiple miRNAs have binding sites, and we confirmed that LINC00691 could bind with miR-1256 through RNA pull down experiments. Then, we found that miR-1256 could regulate the protein expression of ST5 through bioinformatics and luciferase reporter gene detection analyses. Western blot analysis showed that LINC00691 could regulate ST5 expression and that LINC00691 regulated ST5 protein expression in a miR-1256-dependent manner. In this study, we verified the regulatory effect of the LINC00691/miR-1256/ST5 signaling pathway on osteosarcoma cells.
ST5, also known as dennd2b, was originally identified by screening a cDNA expression library of gene products that inhibit the tumorigenicity of HeLa cells in nude mice. ST5 is a differentially expressed regulatory gene in the HeLa/fibroblast somatic hybridization system.36 ST5 activates the MAPK signaling pathway downstream of EGFR and binds to the adaptor protein Grb2, which contains the SH3 domain, and Grb2 mediates the transport of EGFR.37 Proteins containing the denn domain are regulated by phosphorylation; for example, AKT phosphorylation of dennd1a relieves its self-inhibition and promotes GEF activity.38 However, the regulatory role of ST5 in osteosarcoma cells has not been reported. We found that ST5, as a tumor suppressor gene, regulates the function of osteosarcoma cells.
In this study, we verified the regulatory mechanism by which the LINC00691/miR-1256/ST5 signaling pathway fine-tunes the function of osteosarcoma cells. The LINC00691/miR-1256/ST5 signaling pathway inhibited the proliferation, migration, and invasion of osteosarcoma cells and promoted G1 phase arrest of osteosarcoma cells. LINC00691 inhibits the invasion of osteosarcoma cells by inhibiting EMT. This study will provide new ideas and targets for the treatment of osteosarcoma.
Acknowledgments
This work was supported by funds from the China Postdoctoral Science Foundation (2019M661633).
This work was supported by funds from the Science and Technology Commission of Shanghai Municipality (19441902700, 18441903100).
This work was supported by funds from the Shandong Key Research and Development Plan (2019GSF107057) and the Shandong Medical and Health Science and Technology Development Program (No. 2017WS446).
Disclosure
The authors report no conflicts of interest for this work.
References
1. Ritter J, Bielack SS. Osteosarcoma. Ann Oncol. 2010;21(Suppl 7):vii320–vii325. doi:10.1093/annonc/mdq276
2. Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82:690–698.
3. Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195. doi:10.1016/j.canlet.2016.11.019
4. Kansara M, Teng MW, Smyth MJ, Thomas DM. Translational biology of osteosarcoma. Nat Rev Cancer. 2014;14:722–735.
5. Ferrari S, Serra M. An update on chemotherapy for osteosarcoma. Expert Opin Pharmacother. 2015;16:2727–2736. doi:10.1517/14656566.2015.1102226
6. Wedekind MF, Wagner LM, Cripe TP. Immunotherapy for osteosarcoma: where do we go from here? Pediatr Blood Cancer. 2018;65:e27227.
7. Wycislo KL, Fan TM. The immunotherapy of canine osteosarcoma: a historical and systematic review. J Vet Intern Med. 2015;29:759–769. doi:10.1111/jvim.12603
8. Wang JY, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627. doi:10.1016/j.biopha.2019.109627
9. Li YH, Hu YQ, Wang SC, Li Y, Chen DM. LncRNA SNHG5: a new budding star in human cancers. Gene. 2020;749:144724. doi:10.1016/j.gene.2020.144724
10. Zhou Y, Tian B, Tang J, et al. SNHG7: a novel vital oncogenic lncRNA in human cancers. Biomed Pharmacother. 2020;124:109921. doi:10.1016/j.biopha.2020.109921
11. Zhang Y, Pu Y, Wang J, Li Z, Wang H. Research progress regarding the role of long non-coding RNAs in osteosarcoma. Oncol Lett. 2020;20:2606–2612. doi:10.3892/ol.2020.11807
12. Yang X, Xie Z, Lei X, Gan R. Long non-coding RNA GAS5 in human cancer. Oncol Lett. 2020;20:2587–2594. doi:10.3892/ol.2020.11809
13. Tang F, Wang H, Chen E, et al. LncRNA-ATB promotes TGF-beta-induced glioma cells invasion through NF-kappaB and P38/MAPK pathway. J Cell Physiol. 2019;234:23302–23314. doi:10.1002/jcp.28898
14. Ye Y, Shen A, Liu A. Long non-coding RNA H19 and cancer: a competing endogenous RNA. Bull Cancer. 2019;106:1152–1159. doi:10.1016/j.bulcan.2019.08.011
15. Zhao Y, Feng C, Li Y, Ma Y, Cai R. LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem. 2019;460:1–8. doi:10.1007/s11010-019-03564-1
16. Zhuang C, Ma Q, Zhuang C, Ye J, Zhang F, Gui Y. LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. FASEB J. 2019;33:11045–11059. doi:10.1096/fj.201900078RR
17. Duan H, Li X, Chen Y, Wang Y, Li Z. LncRNA RHPN1-AS1 promoted cell proliferation, invasion and migration in cervical cancer via the modulation of miR-299-3p/FGF2 axis. Life Sci. 2019;239:116856. doi:10.1016/j.lfs.2019.116856
18. Xiu B, Chi Y, Liu L, et al. LINC02273 drives breast cancer metastasis by epigenetically increasing AGR2 transcription. Mol Cancer. 2019;18:187. doi:10.1186/s12943-019-1115-y
19. Chen F, Li Z, Deng C, Yan H. Integration analysis for novel lncRNA markers predicting tumor recurrence in human colon adenocarcinoma. J Transl Med. 2019;17:299. doi:10.1186/s12967-019-2049-2
20. Deng W, Zhang Y, Cai J, et al. LncRNA-ANRIL promotes gastric cancer progression by enhancing NF-kB signaling. Exp Biol Med (Maywood). 2019;244:953–959. doi:10.1177/1535370219860207
21. Chi Y, Wang J, Wang J, Yu W, Yang J. Long non-coding RNA in the pathogenesis of cancers. Cells. 2019;8:1015. doi:10.3390/cells8091015
22. Erickson CA. The deformability of BHK cells and polyoma virus-transformed BHK cells in relation to locomotory behaviour. J Cell Sci. 1980;44:187–200.
23. Chen W, Chen M, Xu Y, et al. Long non-coding RNA THOR promotes human osteosarcoma cell growth in vitro and in vivo. Biochem Biophys Res Commun. 2018;499:913–919. doi:10.1016/j.bbrc.2018.04.019
24. Wang B, Qu XL, Liu J, Lu J, Zhou ZY. HOTAIR promotes osteosarcoma development by sponging miR-217 and targeting ZEB1. J Cell Physiol. 2019;234:6173–6181. doi:10.1002/jcp.27394
25. Kotake Y, Goto T, Naemura M, Inoue Y, Okamoto H, Tahara K. Long noncoding RNA PANDA positively regulates proliferation of osteosarcoma cells. Anticancer Res. 2017;37:81–85. doi:10.21873/anticanres.11292
26. Zhao W, Qin P, Zhang D, et al. Long non-coding RNA PVT1 encapsulated in bone marrow mesenchymal stem cell-derived exosomes promotes osteosarcoma growth and metastasis by stabilizing ERG and sponging miR-183-5p. Aging. 2019;11:9581–9596. doi:10.18632/aging.102406
27. Cong M, Jing R. Long non-coding RNA TUSC7 suppresses osteosarcoma by targeting miR-211. Biosci Rep. 2019;39. doi:10.1042/BSR20190291
28. Bi X, Guo XH, Mo BY, et al. LncRNA PICSAR promotes cell proliferation, migration and invasion of fibroblast-like synoviocytes by sponging miRNA-4701-5p in rheumatoid arthritis. EBioMedicine. 2019;50:408–420. doi:10.1016/j.ebiom.2019.11.024
29. Li D, Zhang J, Li J. Role of miRNA sponges in hepatocellular carcinoma. Clin Chim Acta. 2020;500:10–19.
30. Dhawan A, Harris AL, Buffa FM, Scott JG. Endogenous miRNA sponges mediate the generation of oscillatory dynamics for a non-coding RNA network. J Theor Biol. 2019;481:54–60. doi:10.1016/j.jtbi.2018.10.055
31. Militello G, Weirick T, John D, Doring C, Dimmeler S, Uchida S. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18:780–788.
32. Zhu C, Huang L, Xu F, Li P, Li P, Hu F. LncRNA PCAT6 promotes tumor progression in osteosarcoma via activation of TGF-beta pathway by sponging miR-185-5p. Biochem Biophys Res Commun. 2019.
33. Lin H, Zhao Z, Hao Y, He J, He J. 1Long noncoding RNA HIF1A-AS2 facilitates cell survival and migration by sponging miR-33b-5p to modulate SIRT6 expression in osteosarcoma. Biochem Cell Biol. 2019.
34. Luo W, He H, Xiao W, et al. MALAT1 promotes osteosarcoma development by targeting TGFA via MIR376A. Oncotarget. 2016;7:54733–54743. doi:10.18632/oncotarget.10752
35. Wang Y, Kong D. Knockdown of lncRNA MEG3 inhibits viability, migration, and invasion and promotes apoptosis by sponging miR-127 in osteosarcoma cell. J Cell Biochem. 2018;119:669–679. doi:10.1002/jcb.26230
36. Oshimura M, Kugoh H, Koi M, et al. Transfer of a normal human chromosome 11 suppresses tumorigenicity of some but not all tumor cell lines. J Cell Biochem. 1990;42:135–142. doi:10.1002/jcb.240420304
37. Ioannou MS, Kulasekaran G, Fotouhi M, et al. Intersectin-s interaction with DENND2B facilitates recycling of epidermal growth factor receptor. EMBO Rep. 2017;18:2119–2130. doi:10.15252/embr.201744034
38. Kulasekaran G, Nossova N, Marat AL, et al. Phosphorylation-dependent regulation of connecdenn/DENND1 guanine nucleotide exchange factors. J Biol Chem. 2015;290:17999–18008. doi:10.1074/jbc.M115.636712
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.