Back to Journals » Cancer Management and Research » Volume 12
The lncRNA BCYRN1 Functions as an Oncogene in Human Glioma by Downregulating miR-125a-5p in vitro
Authors Yu W, Xiang D, Jia H, He X, Sheng J, Long Y, Zhu S, Wang K, Liu Q
Received 15 August 2019
Accepted for publication 10 January 2020
Published 13 February 2020 Volume 2020:12 Pages 1151—1161
DOI https://doi.org/10.2147/CMAR.S227327
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Wei Yu,1,* Dulei Xiang,1,* Houjun Jia,2,* Xin He,1 Jie Sheng,1 Yuxiang Long,1 Shujuan Zhu,1 Kejian Wang,1 Qian Liu1
1Institute of Neuroscience, Chongqing Medical University, Chongqing 400016, People’s Republic of China; 2Department of Gastrointestinal Surgery, The First Affiliated Hospital of Chongqing Medical University, Chongqing 400016, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qian Liu Tel +86 186 2355 1357
Email [email protected]
Introduction: Numerous studies have demonstrated that long noncoding RNAs (lncRNAs) are deregulated in many cancers and exert their functions through multiple cancer-related biological processes. Glioma is the most common primary malignant central nervous system tumor and has a high fatality rate in adults. In current study, we aimed to determine the role and functional mechanism of the lncRNA BCYRN1 in glioma.
Methods: Gain-of-function and loss-of function approaches were used to investigate the function of BCYRN1. The effects of BCYRN1 on glioma cell proliferation, migration and invasion were evaluated using MTS, Transwell and wound-healing assays. The correlation between the expression of BCYRN1 and miR-125a-5p was verified by quantitative real-time PCR.
Results: The upregulation of BCYRN1 promoted the proliferation, migration and invasion of glioma cells. Meanwhile, the knockdown of BCYRN1 had the opposite effects. BCYRN1 was negatively correlated with miR-125a-5p. Additionally, TAZ, the endogenous target of miR-125a-5p, could be regulated by BCYRN1 in RNA and protein levels. A miR-125a-5p inhibitor restored BCYRN1 siRNA function in glioma.
Conclusion: The present study indicates that BCYRN1 promotes glioma cell proliferation, invasion and migration in vitro. Mechanistically, upregulated expression of BCYRN1 in glioma acts as a sponge to sequester the endogenous tumor suppressor miR-125a-5p and to further increase the expression TAZ. Our findings suggest that BCYRN1 is a novel oncogene and a new therapeutic target for glioma.
Keywords: BCYRN1, glioma, miR-125a-5p, ceRNA, TAZ
Introduction
Gliomas are primary central nervous system tumors that originate from glial cells and are mostly located in the brain. High-grade gliomas tend to metastasize and are usually diagnosed at an advanced stage, rendering curative treatment rare even with a combined approach of surgery, radiation therapy and chemotherapy. The prognosis for patients with malignant gliomas is extremely poor, with a high risk of recurrence.1,2 Therefore, the underlying molecular and cellular mechanisms of glioma pathogenesis must be investigated; additionally, identification of diagnostic biomarkers and potential therapeutic targets is urgently needed.
Recent studies have surprisingly revealed that more than 98% of transcripts do not have protein-coding ability; such transcripts are termed noncoding RNAs (ncRNAs).3 Among them, long noncoding RNAs (lncRNAs), which are defined as a heterogeneous class of transcripts that are longer than 200 nucleotides and do not have protein-coding ability, have attracted considerable attention. Increasing evidence has shown that lncRNAs play a critical role in multiple physiological and pathological biological processes and that dysregulation of lncRNA expression levels is closely related to a wide range of diseases, especially the tumorigenesis and metastasis of multiple cancers, including gliomas.4,5 For instance, CCAT2,6 MALAT1,7 SNHG6,8 UCA1,9 DANCR10 and H1911 function as oncogenes and were overexpressed in gliomas. Meanwhile, TUSC712 and TSLC1-AS113 function as tumor suppressors that were downregulated in gliomas. Although the function of a growing number of lncRNAs in tumors has been well studied, a large number of lncRNAs remain functionally undefined.
LncRNAs exert their functions through multiple mechanisms, including epigenetic silencing, lncRNA-miRNA interactions, lncRNA-protein interactions and lncRNA-mRNA interactions during transcriptional or posttranscriptional processing.5 Additionally, the competitive endogenous RNA (ceRNA) hypothesis has provoked substantial interest; accumulating experimental evidence has illustrated that lncRNAs can act as ceRNAs that share and compete for miRNA response elements (MREs) with target mRNAs. ceRNAs can “sponge” or “decoy” miRNA, decrease the amount of available miRNAs and contribute to the enhanced translation of their target mRNAs.14,15 Previous studies have indicated that the lncRNA SNHG12 can modulate the expression of Notch2 by sponging miRNA in osteosarcoma.16 Furthermore, in gastric cancer, linc00483 enhances the expression of SPAG9, which can activate the MAPK signaling pathway by competitively sequestering miR-30a-3p and promoting gastric cancer cell proliferation and metastasis.17 Emerging evidence suggests that ceRNAs play a critical role in glioma progression. For instance, the lncRNA miR155HG can modulate the expression of ANXA2 by sponging miR-185, which contributes to glioblastoma growth and progression.18 Furthermore, LINC00689 functions as a ceRNA by directly interacting with miR-338-3p to upregulate pyruvate kinase M2 (PKM2) expression to promote the growth, metastasis and glycolysis of glioma cells.19
Brain cytoplasmic RNA 1 (BCYRN1), also called BC200, is selectively expressed in the central nervous system.20 Recent studies have found that BCYRN1 is more highly expressed in breast, ovarian, colon, cervical and other cancer tissues than in corresponding normal tissues, and BCYRN1 is related to the tumorigenesis and prognosis of these cancers.21,22 Our previous studies have shown that BCYRN1 is significantly downregulated during genotoxic stress-induced necrosis in U87 and U251 cells, indicating that BCYRN1 may have oncogenic potential in glioma cells.23 In the present study, we report that BCYRN1 can promote glioma cell proliferation, invasion and migration in vitro. By acting as a sponge to sequester the endogenous tumor suppressor miR-125a-5p, BCYRN1 further increases the expression of TAZ.
Materials and Methods
Cell Culture and Transfection
The glioma cell lines U251 and U87 were obtained from the Chinese Academy of Sciences Cell Bank (Shanghai). The cells were grown in DMEM (HyClone, USA) supplemented with 10% fetal bovine serum (HyClone, USA), 1% penicillin-streptomycin and 1% L-glutamine and maintained in a humidified atmosphere of 5% CO2 in an incubator at 37°C. U251 and U87 cells were transfected with plasmids using the transfection reagent Lipofectamine 3000 (Invitrogen, USA) or with oligonucleotides by Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions.
Plasmid Construction and Oligonucleotides
The complementary DNA coding for BCYRN1 was PCR amplified by EX Taq (TaKaRa, Japan) and subcloned into MSCV. The primers used were 5ʹ- ATTCTAGAGCTAGCGAATTCGGCCGGGCGCGGTGGCTCAC −3ʹ (forward) and 5ʹ-GAGCGATCGCAGATCCTTGCGGCCGCAAAGGGGGGGGGGGGTTGTT −3ʹ (reverse). All plasmids were sent to Invitrogen(USA) for sequencing. Small interfering RNA (siRNA) targeting BCYRN1, a miR-125a-5p mimic, miR-125a-5p inhibitors and a siRNA negative control (si-NC) were purchased from GenePharma (Shanghai). All of the oligonucleotide sequences are listed in Table 1.
 |
Table 1 The Primers and siRNA Sequences Used in This Study |
Cell Proliferation Assay
Cell proliferation was assessed using 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay (Promega, USA) according to the manufacturer’s protocol. The absorbance was measured at 490 nm on a microplate reader (Thermo Scientific, USA).
Wound-Healing Assay
Cells were cultured in twelve-well plates and grown to confluence overnight. Linear wounds were made with a sterile pipette tip. Cells were washed with PBS to remove debris and incubated at 37°C for 24 h. Images showing cell migration at 0 h and 24 h after scratching were obtained using an inverted microscope (Nikon, Japan) at a magnification of 200×.
Transwell Assays
For the invasion assay, 2×104 cells were plated in the top transwell chamber (24-well insert; 8-μm pore size, BD Biosciences, USA) with Matrigel matrix (Sigma, USA) according to the manufacturer’s instructions. For the migration assay, transwell chambers without Matrigel were used. In both assays, cells were plated in medium without serum, and medium supplemented with 10% serum was used as a chemo-attractant in the lower chamber. The cells were incubated for 24 h. Cells that did not migrate or invade through the pores were removed by a cotton swab. Filters were fixed with 90% ethanol, stained with 0.1% crystal violet, and photographed. Cell numbers were counted.
Quantitative Real-Time PCR (qPCR)
Total RNA was isolated using Trizol reagent (TaKaRa, Japan). The cDNA of miRNA was generated using the Mir-X™ miRNA First-Strand Synthesis Kit (Clontech, Japan). The cDNA of lncRNA was generated using the PrimeScript™ RT Reagent Kit (TaKaRa, Japan). qPCR was performed in triplicate using SYBR Premix Ex Taq II (TaKaRa, Japan) with a CFX96 Real-Time PCR Detection System (Bio-Rad, USA). The relative RNA expression levels were calculated by the Vandesompele method. ACTB and GAPDH were used as endogenous controls for lncRNA, as were U6 and RNU for miRNA. The primer sequences used in qPCR are listed in Table 1.
Western Blot Analysis
Total protein was extracted from transfected cells using the mammalian protein extraction reagent RIPA (Beyotime, China). Total proteins from the lysates were separated on a 10% SDS-PAGE gel and then transferred to PVDF membranes (Millipore, USA). The membranes were blocked in 5% nonfat milk and then incubated with diluted specific primary antibodies against TAZ (Proteintech, USA) and ATCB (Proteintech, USA) overnight at 4°C. Primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies. The bands were visualized using an enhanced chemiluminescent kit (ECL kit, Santa Cruz, USA) according to the manufacturer’s protocol.
Statistical Analysis
Student’s t-test was used for comparisons. SPSS 20.0 software was used for analysis. Differences were defined as statistically significant at p-values < 0.05.
Results
Silencing BCYRN1 Inhibits the Proliferation and Metastasis of Glioma
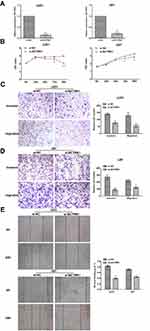
To evaluate the biological functions of BCYRN1 in glioma in vitro, we silenced BCYRN1 in two glioma cell lines, U251 and U87, by transfection of the specific siRNA si-BCYRN1 and using si-NC as a negative control (Figure 1A). As demonstrated by the MTS assay, repression of BCYRN1 significantly decreased the proliferation of U251 and U87 cells (Figure 1B). Furthermore, the transwell assays showed that silencing BCYRN1 suppressed glioma cell migration and invasion (Figure 1C and D). A wound-healing assay suggested that the migratory ability of U87 and U251 cells was obviously decreased after BCYRN1 knockdown (Figure 1E).
Overexpression of BCYRN1 Promotes the Proliferation and Metastasis of Glioma
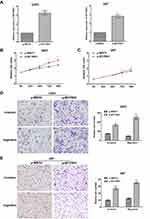
To further confirm the role of BCYRN1 in glioma progression, we enhanced BCYRN1 expression by transfecting a BCYRN1 expression vector (p-BCYRN1) into the glioma cell lines U251 and U87 and using the p-MSCV vector as a negative control (Figure 2A). MTS assays indicated that the enhanced expression of BCYRN1 significantly increased the proliferation of U251 and U87 cells (Figure 2B and C). Next, we investigated the impact of BCYRN1 overexpression on glioma cell invasion and migration. Transwell assays indicated that the number of invading cells in the p-BCYRN1 group was largely increased compared to that in the p-MSCV group (Figure 2D and E).
BCYRN1 Binds to miR-125a-5p and Negatively Regulates Its Expression
Many lncRNAs have recently been reported to function as ceRNAs by competitively binding to microRNAs. To further understand the underlying mechanism of BCYRN1 in glioma, we performed a bioinformatic analysis using LncBase Predicted V.2 and found that BCYRN1 has a putative binding site on miR-125a-5p (Figure 3A). Then, through overexpression and knockdown of BCYRN1, we found that BCYRN1 had an inverse correlation with miR-125a-5p in U251 cells (Figure 3B and C). Additionally, through overexpression and knockdown of miR-125a-5p, the same negative relationship was found between BCYRN1 and miR-125a-5p (Figure 3D and E). These findings suggested that miR-125a-5p interacts with BCYRN1.
BCYRN1 Modulates the Expression of the Endogenous miR-125a-5p Target TAZ
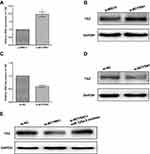
Previous evidence has demonstrated that miR-125a-5p can directly target the 3ʹ UTR of TAZ in glioma cells. Thus, we assumed that BCYRN1 may also function in this pathway. We found that overexpression of BCYRN1 can upregulate the mRNA and protein levels of TAZ(Figure 4A and B) and that knockdown of BCYRN1 can downregulate the mRNA and protein levels of TAZ (Figure 4C and D). TAZ expression can be downregulated by knockdown of BCYRN1, which can be reversed by a miR-125a-5p inhibitor (Figure 4E).
miR-125a-5p Inhibition Restores BCYRN1 siRNA Function
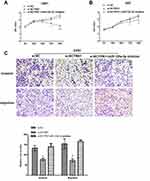
To determine the effects of BCYRN1 and miR-125a-5p on the biological functions of glioma cells, we transfected glioma cells with the following combinations: si-NC, si-BCYRN1, or si-BCYRN1 and miR-125a-5p inhibitors. The results showed that si-BCYRN1 dramatically suppressed cell proliferation, invasion and migration compared with the negative control group, and miR-125a-5p inhibition rescued these inhibitory effects of BCYRN1 (Figure 5A–C).
Discussion
Gliomas are the most common and devastatingly invasive tumors of the central nervous system and have fatal outcomes. Over the past several decades, emerging studies have illustrated that altered expression of lncRNAs contributes to cancer pathogenesis, thus providing new insights into the diagnosis and treatment of cancers. In the present study, the biological function of BCYRN1 in glioma was explored for the first time, and we uncovered its potential role as a ceRNA that sponges miR-125a-5p.
In our previous study, we have showed that BCYRN1 is significantly downregulated in glioma cells treated with high concentrations of DOX, which can induce genotoxic stress-induced necrosis of cells.23 Therefore, we speculated that BCYRN1 may have oncogenic potential. However, Kraus et al investigated 90 lncRNAs in 30 tissue specimens, including 3 different glioma entities and normal brain tissues, and found that 4 lncRNAs, including BCYRN1, are stably expressed in the three glioma entities and normal brain tissues.24 Thus, the role of BCYRN1 in glioma remains controversial. In this study, we characterized the function of BCYRN1 in glioma cells by applying gain-of-function and loss-of-function approaches. Overexpression of BCYRN1 promoted the proliferation, migration and invasion of glioma cells. Meanwhile, knockdown of BCYRN1 had the opposite effects. Therefore, our results indicated that BCYRN1 functions as an oncogene in glioma. These data are consistent with previous reports indicating that BCYRN1 also acts as an oncogene in other cancers, such as cervical and colon cancer.21,22
The ceRNA hypothesis has provided a network regulatory model for ceRNA-miRNA-mRNA interactions, and emerging data have revealed that lncRNAs can act as ceRNAs to regulate biological processes in many cancers.14,15,25,26 Interestingly, Peng et al reported that decreasing BCYRN1 expression attenuates the viability and motility of cervical cancer cells through targeting of miR-138.22 Additionally, many lncRNAs, such as MALAT1,27 MNX1-AS1,28 LINC0015229,30 and DANCR,31 have been verified to act as ceRNAs that regulate the tumorigenesis and progression of gliomas. Inspired by the ceRNA regulatory network, we hypothesized that BCYRN1 may also serve as a ceRNA by directly binding to specific miRNAs. Subsequently, we searched for potential interactions with miRNAs using bioinformatics databases. Among several potential miRNAs, miR-125a-5p attracted our interest, and a putative binding site between BCYRN1 and mi-125a-5p was predicted using LncBase Predicted V.2. MiR-125a-5p is derived from the 5ʹ end of pre-miR-125a.32 Recently, miR-125a-5p has demonstrated to be downregulated in glioma, to suppress the proliferation of glioma and to promote the cell differentiation of glioma by directly targeting the 3ʹ UTR of TAZ.33 TAZ is an important transducer of the Hippo tumor-suppressor pathway, and knockdown of TAZ in glioma stem cells decreases the expression of mesenchymal markers, as well as invasion, self-renewal and tumor formation.34 Thus, we hypothesized that BCYRN1 may function in glioma by modulating TAZ by interacting with miR-125a-5p. In this study, we found that BCYRN1 can reduce miR-125a-5p expression and that miR-125a-5p can downregulate BCYRN1 expression, indicating that BCYRN1 and miR-125a-5p interact and can degrade each other. Furthermore, we confirmed that BCYRN1 can decrease the expression of TAZ at both the mRNA and protein levels, whereas miR-125a-5p inhibitors can partially reverse the reduction caused by BCYRN1 knockdown. Additionally, we found that the inhibition of proliferation and metastasis due to BCYRN1 knockdown can also be partially reversed by miR-125a-5p inhibition. These data imply that BCYRN1 may exert its function through the BCYRN1-miR-125a-5p-TAZ axis.
However, in the present study, we did not prove that BCYRN1 and miR-125a-5p directly interact, and we only investigated one miRNA and lncRNA pair. The ceRNA activity of BCYRN1 may be able to sponge several miRNAs at once, while miR-125a-5p is also capable of controlling other genes. We speculate that other ceRNAs may regulate the expression of key genes in glioma.
Collectively, this is the first report that BCYRN1 served as an oncogene to promote the proliferation, invasion and migration of glioma cells. Furthermore, our results also indicated that BCYRN1 may be a molecular sponge for miR-125a-5p, leading to upregulation of its endogenous target, TAZ. Further studies are required to explore the potential mechanism of BCYRN1. Our results showed that BCYRN1 may be a biomarker for diagnosis and a promising therapeutic target in glioma.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (Project #81502161) and the Chongqing Science and Technology Commission (Project #cstc2015jcyjA10007).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Pace A, Dirven L, Koekkoek JAF, et al. European Association for Neuro-Oncology (EANO) guidelines for palliative care in adults with glioma. Lancet Oncol. 2017;18(6):e330–e340. doi:10.1016/S1470-2045(17)30345-5
2. Sizoo EM, Pasman HR, Dirven L, et al. The end-of-life phase of high-grade glioma patients: a systematic review. Support Care Cancer. 2014;22(3):847–857. doi:10.1007/s00520-013-2088-9
3. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74.
4. Leucci E. Cancer development and therapy resistance: spotlights on the dark side of the genome. Pharmacol Ther. 2018;189:22–30. doi:10.1016/j.pharmthera.2018.04.001
5. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
6. Zeng J, Du T, Song Y, et al. Knockdown of long noncoding RNA CCAT2 inhibits cellular proliferation, invasion, and epithelial-mesenchymal transition in glioma cells. Oncol Res. 2017;25(6):913–921. doi:10.3727/096504016X14792098307036
7. Xiong Z, Wang L, Wang Q. LncRNA MALAT1/miR-129 axis promotes glioma tumorigenesis by targeting SOX2. J Cell Mol Med. 2018;22:3929–3940. doi:10.1111/jcmm.2018.22.issue-8
8. Meng Q, Yang BY, Liu B, Yang JX, Sun Y. Long non-coding RNA SNHG6 promotes glioma tumorigenesis by sponging miR-101-3p. Int J Biol Markers. 2018;33(2):148–155. doi:10.1177/1724600817747524
9. He Z, You C, Zhao D. Long non-coding RNA UCA1/miR-182/PFKFB2 axis modulates glioblastoma-associated stromal cells-mediated glycolysis and invasion of glioma cells. Biochem Biophys Res Commun. 2018;500(3):569–576. doi:10.1016/j.bbrc.2018.04.091
10. Li J, Zhou L. Overexpression of lncRNA DANCR positively affects progression of glioma via activating Wnt/beta-catenin signaling. Biomed Pharmacother. 2018;102:602–607. doi:10.1016/j.biopha.2018.03.116
11. Chen L, Wang Y, He J, Zhang C, Chen J, Shi D. Long non-coding RNA H19 promotes proliferation and invasion in human glioma cells by downregulating miR-152. Oncol Res. 2018. doi:10.3727/096504018X15178768577951
12. Shang C, Guo Y, Hong Y, Xue YX. Long non-coding RNA TUSC7, a target of miR-23b, plays tumor-suppressing roles in human gliomas. Front Cell Neurosci. 2016;10:235. doi:10.3389/fncel.2016.00235
13. Qin X, Yao J, Geng P, Fu X, Xue J, Zhang Z. LncRNA TSLC1-AS1 is a novel tumor suppressor in glioma. Int J Clin Exp Pathol. 2014;7(6):3065–3072.
14. Smillie CL, Sirey T. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53(3):231–245. doi:10.1080/10409238.2018.1447542
15. Deng K, Wang H, Guo X, Xia J. The cross talk between long, non-coding RNAs and microRNAs in gastric cancer. Acta Biochim Biophys Sin (Shanghai). 2016;48(2):111–116. doi:10.1093/abbs/gmv120
16. Zhou S, Yu L, Xiong M, Dai G. LncRNA SNHG12 promotes tumorigenesis and metastasis in osteosarcoma by upregulating Notch2 by sponging miR-195-5p. Biochem Biophys Res Commun. 2018;495(2):1822–1832. doi:10.1016/j.bbrc.2017.12.047
17. Li D, Yang M, Liao A, et al. Linc00483 as ceRNA regulates proliferation and apoptosis through activating MAPKs in gastric cancer. J Cell Mol Med. 2018;22:3875–3886. doi:10.1111/jcmm.2018.22.issue-8
18. Wu W, Yu T, Wu Y, Tian W, Zhang J, Wang Y. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J Exp Clin Cancer Res. 2019;38(1):133. doi:10.1186/s13046-019-1132-0
19. Liu X, Zhu Q, Guo Y, Xiao Z, Hu L, Xu Q. LncRNA LINC00689 promotes the growth, metastasis and glycolysis of glioma cells by targeting miR-338-3p/PKM2 axis. Biomed Pharmacother. 2019;117:109069. doi:10.1016/j.biopha.2019.109069
20. Booy EP, McRae EK, Koul A, Lin F, McKenna SA. The long non-coding RNA BC200 (BCYRN1) is critical for cancer cell survival and proliferation. Mol Cancer. 2017;16(1):109. doi:10.1186/s12943-017-0679-7
21. Wu K, Xu K, Liu K, et al. Long noncoding RNA BC200 regulates cell growth and invasion in colon cancer. Int J Biochem Cell Biol. 2018;99:219–225. doi:10.1016/j.biocel.2018.04.001
22. Peng J, Hou F, Feng J, Xu SX, Meng XY. Long non-coding RNA BCYRN1 promotes the proliferation and metastasis of cervical cancer via targeting microRNA-138 in vitro and in vivo. Oncol Lett. 2018;15(4):5809–5818. doi:10.3892/ol.2018.8015
23. Liu Q, Sun S, Yu W, et al. Altered expression of long non-coding RNAs during genotoxic stress-induced cell death in human glioma cells. J Neurooncol. 2015;122(2):283–292. doi:10.1007/s11060-015-1718-0
24. Kraus TF, Greiner A, Guibourt V, Lisec K, Kretzschmar HA. Identification of Stably Expressed lncRNAs as Valid Endogenous Controls for Profiling of Human Glioma. J Cancer. 2015;6(2):111–119. doi:10.7150/jca.10867
25. Abdollahzadeh R, Daraei A, Mansoori Y, Sepahvand M, Amoli MM, Tavakkoly-Bazzaz J. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–10100. doi:10.1002/jcp.27941
26. Shuwen H, Qing Z, Yan Z, Xi Y. Competitive endogenous RNA in colorectal cancer: a systematic review. Gene. 2018;645:157–162. doi:10.1016/j.gene.2017.12.036
27. Li Z, Xu C, Ding B, Gao M, Wei X, Ji N. Long non-coding RNA MALAT1 promotes proliferation and suppresses apoptosis of glioma cells through derepressing Rap1B by sponging miR-101. J Neurooncol. 2017;134(1):19–28. doi:10.1007/s11060-017-2498-5
28. Gao Y, Xu Y, Wang J, Yang X, Wen L, Feng J. LncRNA MNX1-AS1 promotes glioblastoma progression through inhibition of miR-4443. Oncol Res. 2018.
29. Cai J, Zhang J, Wu P, et al. Blocking LINC00152 suppresses glioblastoma malignancy by impairing mesenchymal phenotype through the miR-612/AKT2/NF-kappaB pathway. J Neurooncol. 2018;140(2):225–236. doi:10.1007/s11060-018-2951-0
30. Chen X, Li D, Gao Y, et al. Long intergenic noncoding RNA 00152 promotes glioma cell proliferation and invasion by interacting with MiR-16. Cell Physiol Biochem. 2018;46(3):1055–1064. doi:10.1159/000488836
31. Xu D, Yu J, Gao G, Lu G, Zhang Y, Ma P. LncRNA DANCR functions as a competing endogenous RNA to regulate RAB1A expression by sponging miR-634 in glioma. Biosci Rep. 2018;38(1). doi:10.1042/BSR20171664
32. Yang X, Qiu J, Kang H, Wang Y, Qian J. miR-125a-5p suppresses colorectal cancer progression by targeting VEGFA. Cancer Manag Res. 2018;10:5839–5853. doi:10.2147/CMAR.S161990
33. Yuan J, Xiao G, Peng G, et al. MiRNA-125a-5p inhibits glioblastoma cell proliferation and promotes cell differentiation by targeting TAZ. Biochem Biophys Res Commun. 2015;457(2):171–176. doi:10.1016/j.bbrc.2014.12.078
34. Bhat KP, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–2609. doi:10.1101/gad.176800.111
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.