Back to Journals » Journal of Inflammation Research » Volume 15
The Influence of Arginine Methylation in Immunity and Inflammation
Authors Srour N, Khan S , Richard S
Received 26 February 2022
Accepted for publication 6 May 2022
Published 13 May 2022 Volume 2022:15 Pages 2939—2958
DOI https://doi.org/10.2147/JIR.S364190
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Nivine Srour,1,2,* Sarah Khan,1,2,* Stephane Richard1,2
1Segal Cancer Centre, Lady Davis Institute for Medical Research, Jewish General Hospital, Montréal, Québec, H3T 1E2, Canada; 2Gerald Bronfman Department of Oncology, and Departments of Biochemistry, Human Genetics, and Medicine, McGill University, Montréal, Québec, H3T 1E2, Canada
*These authors contributed equally to this work
Correspondence: Stephane Richard, Email [email protected]
Abstract: Exploration in the field of epigenetics has revealed that protein arginine methyltransferases (PRMTs) contribute to disease, and this has given way to the development of specific small molecule compounds that inhibit arginine methylation. Protein arginine methylation is known to regulate fundamental cellular processes, such as transcription; pre-mRNA splicing and other RNA processing mechanisms; signal transduction, including the anti-viral response; and cellular metabolism. PRMTs are also implicated in the regulation of physiological processes, including embryonic development, myogenesis, and the immune system. Finally, the dysregulation of PRMTs is apparent in cancer, neurodegeneration, muscular disorders, and during inflammation. Herein, we review the functions of PRMTs in immunity and inflammation. We also discuss recent progress with PRMTs regarding the modulation of gene expression related to T and B lymphocyte differentiation, germinal center dynamics, and anti-viral signaling responses, as well as the clinical relevance of using PRMT inhibitors alone or in combination with other drugs to treat cancer, immune, and inflammatory-related diseases.
Keywords: PRMTs, epigenetics, histones, arginine methylation, immune, inflammation
Arginine Methylation and Inflammation
Arginine methylation is a common post-translational modification in mammalian cells.1 Protein arginine methyltransferases (PRMTs) are the primary enzymes responsible for catalyzing the formation of methylarginines in proteins. PRMTs catalyze the transfer of methyl groups from S-adenosylmethionine (SAM) to the ω-guanidino nitrogen atoms of arginines in proteins.2 There are nine PRMTs with three separate types of activity.3 Type I PRMTs (PRMT1, PRMT2, PRMT3, PRMT4 (herein referred to as CARM1 for co-activator-associated methyltransferase 1), PRMT6, and PRMT8) catalyze the formation of monomethylarginine (MMA) and asymmetric dimethylarginine (aDMA). Type II PRMTs (PRMT5 and PRMT9) catalyze the formation of MMA and symmetric dimethylarginine (sDMA). PRMT7 is the only known type III PRMT, and it catalyzes only the formation of MMA (Figure 1A). There are currently no known dedicated arginine demethylases,3 in contrast to the known family of Jumonji (Jmj) C (JmjC) lysine demethylases (KDMs).4 Therefore, arginine methylation is largely considered to be a long-lasting, as opposed to transient and reversible, post-translational modification. JmjD6 was wrongfully reported as an arginine demethylase as it is a hydroxylase for lysines.5 KDM3A, KDM4E, and KDM5C, known histone methyl lysine demethylases, also possess the ability to demethylate methylarginines in vitro,6 but whether this weak activity is of physiological relevance remains to be shown. The protein arginine deiminase (PAD) family may offer the possibility to reverse methylarginine by converting it to neutral citrulline,7 however, monomethylarginine is a poorer substrate than unmodified arginine.7 Thus, we still await identification of enzymes capable of reversing methylarginine to arginine. Without affecting charge, the addition of methyl groups sterically disrupts hydrogen bonding at affected guanidino nitrogen atoms influencing “reader” association such as Tudor, plant homeodomain (PHD), and WD40 domain-containing proteins.8
PRMTs play a significant role in gene regulation by methylating histone marks.9 PRMT1-catalyzed H4R3me2a and CARM1-catalyzed H3R17me2a, H3R26me2a and H3R42me2a and PRMT5 mediated H3R2me2s are activating histone marks, while PRMT5-catalyzed H2AR3me2s, H4R3me2s, and H3R8me2s; PRMT6-catalyzed H3R2me2a; and CARM1-catalyzed H2AR29me2a are repressive histone marks (Figure 1B). PRMTs also methylate many other substrates to modulate processes and pathways including pre-mRNA splicing, mRNA translation, cell signaling, and DNA damage pathways.3 Many PRMTs favor the methylation of arginine/glycine-rich repeats (RGG/RG motifs) in proteins with the exceptions of CARM1 and PRMT7 that favor arginine/proline-rich repeats (PGM motifs) and RXR motifs, where X is any amino acid, respectively.3
Inflammation is a component of innate immunity, the body’s primary protective response to infection. Pathogenic molecules such as lipopolysaccharides (LPS), double-stranded DNA (dsDNA), and single-stranded RNA (ssRNA) are recognized by pattern recognition receptors (PRRs) such as toll-like receptors (TLRs), retinoic acid-inducible gene I (RIG-I)/melanoma differentiation-associated protein 5 (MDA5), and cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase (cGAS).10 Stimulation of PRRs results in the activation of several signaling pathways, including nuclear factor kappa B (NF-κB), interferon (IFN) regulatory factor (IRF) 3 (IRF3), IRF7, and mitogen-activated protein kinase (MAPK), and subsequently the transcription of genes that encode proinflammatory IFNs and cytokines.10 The NF-κB family consists of five structurally similar proteins (NF-κB1/p50, NF-κB2/p52, RelA/p65, RelB, and c-Rel) that assemble into functional hetero- and homodimers.11 Briefly, canonical activation of the NF-κB pathway involves phosphorylation and activation of the inhibitor of kappa B (IκB) kinase (IKK) complex, followed by phosphorylation, ubiquitination, and degradation of IκB alpha (IκBα) to liberate and allow NF-κB dimers (typically, RelA/p65-NF-κB1/p50 and NF-κB1/p50-c-Rel) to translocate to the nucleus where they can bind to specific κB response elements and stimulate the expression of target genes.11 B and T lymphocytes can also mediate inflammation as part of the body’s adaptive immune system.12 Acute inflammation requires constant stimulation to be maintained, while chronic inflammation arises during sustained inflammation and may lead to autoimmune diseases, such as asthma, systemic lupus erythematosus (SLE), acute graft-versus-host disease (aGVHD), ulcerative colitis, rheumatoid arthritis (RA), and multiple sclerosis (MS) (see below).13–15 It is known that PRMTs are involved in mediating inflammation and, thus, their inhibition may be a promising strategy for the treatment of inflammatory and autoimmune diseases. This review will summarize the currently understood roles of PRMTs in modulating inflammation and the immune response.
PRMT1 in Inflammation
In mammalian cells, PRMT1 is the most active and prevalent type I PRMT.16 It is known to function as a transcriptional co-activator, and it is responsible for the generation of the activation mark, H4R3me2a.17 PRMT1 has also been shown to play roles in the DNA damage response pathway by methylating DNA damage proteins and in RNA metabolism by methylating RNA binding proteins (RBPs) (for review see3). PRMT1 has been recognized as a mediator of inflammation through its interaction with transcription factors and co-activators, including signal transducer and activator of transcription (STAT) proteins, NF-κB, and cyclic adenosine monophosphate (cAMP) response element binding protein (CREB)-binding protein (CBP)/p300-interacting trans-activator 2 (CITED2) (reviewed in18). PRMT1 has also been directly linked to the expression of cytokines and major histocompatibility complex (MHC)-related genes.19 This next section will detail how PRMT1 regulates inflammation.
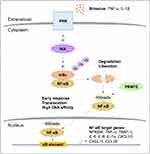
PRMT1 largely functions as a negative regulator of inflammation. Reintjes et al. 2016 showed that PRMT1 directly interacts and methylates the NF-κB subunit, RelA/p65, at R30 to suppress tumor necrosis factor (TNF)-alpha (TNF-α)-induced activation of NF-κB.20 Asymmetric dimethylation of RelA/p65 at R30 interferes with the ability of NF-κB to function as a transcription factor (Figure 2A). Interestingly, depletion of PRMT1 using short hairpin RNA (shRNA) prevented the attenuation of NF-κB target gene expression, normally observed to occur within 4 hours of TNF-α stimulation.20 Further, in response to cytokine interleukin (IL) 4 (IL-4) stimulation, PRMT1 and CARM1 were shown to function as co-activators of STAT5 for the upregulation of CITED2.21 CITED2 negatively regulates NF-κB activation by binding the co-activator CBP/p300 in the nucleus and preventing its association with RelA/p65. This prevents RelA/p65 acetylation, required for its binding and stimulation of target A20 and IL-8 promoters.22 Finally, arginine methylation was shown to play a role in post-transcriptional regulation of MHC-related genes. The use of MTA (5’-methyl-thioadenosine), now known to be a specific inhibitor of PRMT5,23–25 was shown to suppress IFN-γ-induced expression of human leukocyte antigen (HLA) A (HLA-A).26 Moreover, PRMT1 has been linked to the transcriptional repression of hypoxia-inducible factor-1 alpha (HIF-1α) by regulating the activity of transcription factors, specificity protein (Sp) 1 (Sp1) and Sp3.27 Depletion of PRMT1 using small interfering RNA (siRNA) was shown to increase HIF-1α levels and allow CREB to bind to the HLA-B promoter via chromatin remodeling.28 These findings suggest a repressive epigenetic role for PRMT1 in the context of hypoxia, relevant especially to tumor-infiltrating monocytes. Additionally, PRMT1 was shown to methylate class II transactivator (CIITA) to promote its degradation and suppress IFN-γ-induced MHC-II transactivation in macrophages (Figure 2B).19 PRMT1 silencing increased activity at the MHC-II promoter in the presence of IFN-γ and increased expression of HLA-DRA in both primary and transformed mouse peritoneal macrophages.19 These findings provide a function for PRMT1 in vascular inflammation.
Conversely, arginine methylation of the N-terminal domain of nuclear factor of activated T cells (NFAT)-interacting protein (NIP45) by PRMT1, was found to be required for its interaction with NFAT and, thus, the activation of IL-4 and IFN-γ transcription in T helper (Th) 2 (Th2) and Th1 cells, respectively (Figure 2C).29 NIP45 depletion prevents H4R3 methylation and H4 acetylation at relevant promoters, suggesting that PRMT1 can activate inflammation through histone methylation, but still primarily depends on non-histone methylation to initiate the response.30 Significantly, it was shown that NIP45 deletion could ameliorate airway inflammation in asthma by decreasing type 2 innate lymphoid cells (ILC2) differentiation.31 These data suggest that a PRMT1 inhibitor may only be useful to treat inflammation in selected contexts.
PRMT5 in Inflammation
PRMT5 generates the majority of cellular sDMA in mammalian cells. PRMT5 has many substrates (signaling molecules, RNA binding proteins, splicing factors, transcription factors, and histones) to regulate cellular processes (for review see32). Regulation by PRMT5 is critical for transcription; pre-mRNA and alternative splicing; signal transduction; and the DNA damage response (for review see3). In this section, we will discuss how PRMT5 regulates the inflammatory response, particularly through the NF-κB pathway (Figure 3).
PRMT5 was originally cloned as a Janus kinase 2 (JAK2)-binding protein.33 PRMT5 exists in a complex with methylosome protein 50 (MEP50) and a substrate adaptor protein (SAP), including pICln, RIO kinase 1 (RIOK1), and COPR5, to attract and methylate its substrates.34 PRMT5 overwhelmingly serves as a positive regulator of inflammation. In a proteomic screen, PRMT5 was identified as a new TNF-related apoptosis-inducing ligand (TRAIL) receptor-binding protein.35 Interestingly, PRMT5 contributes to TRAIL-induced activation of IKK and NF-κB, thus, leading to the induction of several NF-κB target genes.36 Moreover, it was shown that PRMT5 gene silencing increased TRAIL-mediated cytotoxicity alone without affecting TNF-α-mediated NF-κB signaling. Another study reported that PRMT5-mediated methylation of homeobox A9 (HOXA9), a transcription factor for endothelial cell inflammatory responses, at R140 increased the level of endothelial-leukocyte adhesion molecule (ELAM).37 Depletion of PRMT5 using siRNA led to a loss of E-selectin and vascular cell adhesion protein 1 (VCAM-1) induction, indicating that PRMT5 is an essential component for endothelial cell expression of leukocyte adhesion molecules during the inflammatory response. PRMT5 methylates the RelA/p65 subunit of NF-κB, promoting the expression of the proinflammatory chemokine, C-X-C motif chemokine ligand 10 (CXCL10), in response to TNF-α.38 In addition, PRMT5-mediated methylation of RelA/p65 is required for CXCL11 induction during co-stimulation of endothelial cells with TNF-α and IFN-γ.39 Interestingly, the methylation of RelA/p65 by PRMT5 at R30 increased its DNA binding activity and stimulated the expression of genes encoding cytokines, chemokines, and growth factors, including IL‐1α, IL-8 and TNF receptor‐associated factor 1 (TRAF-1).40 Similar findings also indicate that PRMT5 regulates the NF‐κB signaling through several cell membrane-bound receptors leading to the activation of the IKK complex.41 It was shown that inhibition of PRMT5 methylation diminishes IKKβ and IKKα activation and RelA/p65 nuclear translocation.41 Deletion of PRMT5 using siRNA and its pharmacological inhibition using EPZ015666 were shown to decrease the production of IL‐6 and IL‐8 and prevent cell proliferation, migration, and invasion by attenuating the activation of NF‐κB.41 Recently, PRMT5 was shown to increase VCAM-1 expression via symmetric dimethylation of RelA/p65 on R30.42 PRMT5 knockdown in vascular smooth muscle cells (VSMCs) inhibited vascular inflammation and decreased VCAM-1 expression in mice. Together, these findings define a role for PRMT5 in the inflammatory response and suggest that the inhibition of PRMT5 might be an attractive therapeutic approach to attenuate pathological progression of inflammatory-related diseases.
Many different types of PRMT5 inhibitors have been generated. DS-437 was designed to occupy the SAM binding site and part of the substrate binding pocket of PRMT5 by adding a urea moiety that mimics the guanidinium group of substrate arginines to S-adenosylhomocysteine (SAH). Indeed, DS-437 prevented the methylation of histone H4 by PRMT5 but also was able to inhibit PRMT7; therefore, it is not specific for PRMT5.43 EPZ015666 and GSK3203591 were designed as substrate competitive inhibitors of PRMT5.44,45 These compounds are high-affinity inhibitors of SAM-bound PRMT5 complexes. As MTA was shown to be elevated in MTA phosphorylase (MTAP) negative cancers and to have preference for binding PRMT5,23–25 a new specific inhibitor (MRTX1719) was generated that specifically inhibits the MTA-bound PRMT5-MEP50 complex.46 Other strategies to inhibit PRMT5 include the development of a proteolysis-targeting chimeric (PROTAC) probe (MS4322) to degrade PRMT5.47 Finally, another strategy has been to target the PRMT5 substrate adaptor interaction; BRD0639 disrupts the PRMT5-RIOK1 interactions required for the methylation of a variety of RIOK1-mediated, PRMT5-specific substrates.48 The inhibitors referenced herein are listed in Table 1 (see below).
 |  |  |
Table 1 Defining Immune Function with PRMT Inhibitors |
Type I PRMTs, PRMT6 and CARM1, also positively regulate inflammation. PRMT6 activates NF-κB by directly binding to RelA/p65 and promoting its nuclear translocation.49 Using a gain-of-function PRMT6 allele in mice, it was shown that PRMT6 binds NF-κB-regulated promoters, such as IL-6, and stimulates gene expression upon TNF-α stimulation. CARM1 was also shown to interact directly with RelA/p65 and function as a co-activator at NF-κB target genes in response to TNF-α and LPS stimulation.50 Further, CARM1 was found to participate in NF-κB-mediated transcription by remodeling chromatin via H3R17 methylation at inflammatory gene promoters such as TNF-α, IL-8, and CXCL10 in monocytes.51 Thus, given the prominent role of PRMTs in inflammation, it will be worthwhile to investigate whether certain PRMT inhibitors might enhance the effects of anti-inflammatory drugs.
Physiological Role of PRMTs in the Immune System
Arginine methylation is a major contributor to immune development and function. Several PRMTs were shown to play a critical role in the establishment and maintenance of lymphoid and myeloid cell lines.1,52 In the following section, the main functions of PRMTs in regulating the immune system will be discussed.
PRMT1 is known to affect T lymphocyte function. With the huge success of cancer immunotherapies and the generation of chimeric antigen receptor (CAR)-T cells, it becomes important to further understand the function of PRMT1 in T cells. Interestingly, PRMT1 was found to regulate the Th17 differentiation process.53 PRMT1 interacts with growth factor independent 1 (GFI1), a transcriptional regulator required for development and maintenance of T lymphoid leukemia, to regulate the DNA damage response.54 Moreover, PRMT1 is required for cytokine production by Th cells.55
PRMT1 is also implicated in B lymphocyte regulation. In B cells, PRMT1 methylates cyclin-dependent kinase 4 (CDK4) and thereby prevents the formation of the CDK4-Cyclin-D3 complex and cell cycle progression. This methylation event blocks pre-B-cell proliferation and activates light chain immunoglobulin (IgL) gene assembly and pre-B-cell differentiation.56 Furthermore, PRMT1 is necessary for lymphocyte development, proliferation, and differentiation in vivo.57 PRMT1-deficient mice exhibit defects in B-cell development with diminished levels of serum antibodies by impairing T cell-independent antibody production. Arginine methylation of the Igα subunit of the B cell receptor (BCR) negatively regulates the calcium (Ca2+) and the phosphatidylinositol 3-kinase (PI3K) signaling pathways of the BCR while promoting B cell differentiation.58 PRMT1 deletion in mature B cells also results in reduced B cell activation and differentiation, thereby impairing humoral immunity in vivo.59 Additionally, PRMT1 plays a critical role in IL-6 production in macrophages.60
CARM1 methylates the thymocyte cyclic AMP-regulated phosphoprotein (TARPP), a T cell-specific factor, at R650 and, thus, regulates the differentiation of early thymocyte progenitors.61 Knockout of CARM1 reduces the cluster of differentiation (CD) 4 negative (CD4−) CD8− T cell population in mice and blocks thymocyte developmental at the CD44+ CD25− stage.61,62 Moreover, inhibition of CARM1 in T cells greatly increases CD8+ T cell accumulation in tumors and enhances antitumorgenicity.63 Further, transcriptomic data shows that CARM1 deletion upregulates the expression of TCF7 and MYB, key genes required for the maintenance and self-renewal of memory T cell populations.64,65 CARM1 is also required for Th17 differentiation by opening chromatin at critical gene loci.66 Precisely, CARM1 was shown to generate the activating mark, H3R17me2a, and prevent the deposition of the repressive mark, H3K9me3, at the IL-17 locus, thus, leading to amplified IL-17A transcription and activation of the Th17 differentiation program.66
Little is known about the role of CARM1 during B cell development. However, a recent study has shown that CARM1 is implicated in lineage differentiation for both B and T cells.67 LPS stimulation was found to increase CARM1 expression in B and T lymphocytes as well as monocytes that mediate caspase-3-dependent lymphocyte cell death.67 Inhibition of CARM1 activity using TP-064 attenuated lymphocyte cell death and protected mice following LPS lung injury and polymicrobial sepsis.67 Moreover, CARM1 was shown to downregulate microRNA (miRNA) 223 (miR-223) expression via the methylation of runt-related transcription factor 1 (RUNX1) at residue R223 and lead to the recruitment of double PHD finger 2 (DPF2) to repress myeloid differentiation.68 Strikingly, CARM1 overexpression inhibited the differentiation of adult hematopoietic stem cells (HSCs) in culture, while CARM1 knockdown promoted their differentiation.68 In 2018, Nimer et al. also showed that CARM1 plays a critical role in myeloid leukemogenesis.69 CARM1 depletion minimally impacted normal hematopoiesis but strongly impaired leukemogenesis by disrupting cell cycle progression, promoting myeloid differentiation, and inducing apoptosis.69 Thus, CARM1 could be a potential therapeutic target for certain hematopoietic cancers.
PRMT5-mediated arginine methylation in activated T cells has been shown to be essential for the recruitment of transcription factors during cytokine gene expression. Depletion of PRMT5 in T lymphocytes impairs IL-2 gene expression.70 Moreover, PRMT5 modulates T cell activation processes via the regulation of the transcription of IFN-induced cytokine genes,71 and when deleted, PRMT5 decreases signaling via the γc-family of cytokines and reduces peripheral CD4+ and CD8+ T cell populations.72 PRMT5 depletion in the CD4+ Th cell compartment suppressed Th17 differentiation and protected mice from Th17-mediated diseases such as experimental autoimmune encephalomyelitis (EAE), a mouse model of autoimmune inflammatory diseases of the central nervous system (CNS) (see below).73
PRMT5 was shown to be essential for T cell survival and proliferation by maintaining cytokine signaling.36 PRMT5 expression was also shown to be upregulated during human T cell leukemia virus type-1 (HTLV-1)-mediated T cell transformation, and its inhibition resulted in increased viral gene expression and decreased cellular proliferation.74 Little is known about the role of PRMT5 in the physiological function of B lymphocytes, but it has been shown that PRMT5 mRNA levels, together with protein sDMA levels, are elevated in activated B cells.75 Additionally, PRMT5 is markedly overexpressed in primary Epstein-Barr virus (EBV) lymphomas and lymphoblastoid cell lines.76 Together, these data suggest that PRMT5 overexpression could be a marker of B cellular transformation.76 It was shown that PRMT5 acts in a chromatin-wide repressive manner during B cell transformation via H4R3me2s and H3R8me2s.77,78 Thus, it is likely that inhibiting PRMT5 would block the initiation and maintenance of EBV-driven B lymphocyte transformation and survival without affecting resting and activated B cells. Furthermore, deleting PRMT5 in all hematopoietic cells reduces pro- and pre-B cell differentiation and impairs T cell development, followed by defects in cytokine signaling,79 suggesting that PRMT5 is required for B cell development in the bone marrow. PRMT5 was shown to play an important role in the regulation of antibody responses and germinal center (GC) dynamics during the development of B cells.80 Further understanding of PRMT5 substrates following B and T cell activation is required to define the function of arginine methylation in regulating signaling during lymphopoiesis.
PRMT7 is an essential contributor to B cell lymphomagenesis. While PRMT7 B cell knockout mice survive into adulthood, the loss of PRMT7 reduces mature marginal zone B cell populations and increases native follicular B cell populations, thus, promoting GC formation and plasma cell differentiation.81 Mechanistically, PRMT7 was shown to influence H4R3me2s at the Bcl6 promoter and negatively regulate Bcl6 expression.81 Furthermore, PRMT7-deficient B cell mice secrete low levels of immunoglobulins, IgG1 and IgA.81 Although H4R3me2s is not a histone mark catalyzed by PRMT7, it was shown that PRMT7 monomethylates neighboring H4R17 to allosterically influence PRMT5-mediated H4R3me2s.82 These findings suggest that PRMT7 mediated histone methylation may play a role in the onset and progression of B cell lymphomas. Further studies are required to fully understand the intersection of the activities of different PRMTs in the regulation of B cell development.
Collectively, these findings provide insight into the essential contribution of arginine methylation to B cell and T cell development and provide rationale for targeting PRMTs in different immune cell-related diseases, for example, B cell non-Hodgkin lymphomas. Although PRMTs were linked to malignant B cell survival and proliferation, the overall relevance of PRMT overexpression during the transformation process also remains unclear and requires further investigation.
PRMTs in Immune Diseases
Asthma
Several studies link PRMT1 to allergic asthma. PRMT1 is overexpressed in the lung tissue of antigen-induced pulmonary inflammation (AIPI) E3 rat models and mediates eosinophil recruitment into the lungs in response to IL-4 expression.83,84 Raf kinase inhibitor protein (RKIP) and protein inhibitor of activated STAT1 (PIAS1) are reciprocally expressed in epithelial and fibroblast cells and inhibit IL-1β/NF-κB and IL-4/STAT6-mediated PRMT1 expression, respectively.85 More recently, it was shown that PRMT1 regulates processing of asthma-related miRNAs in lung epithelial cells.86 PRMT1 is recruited, in complex with either STAT1 or RUNX1, to promote processing of miRNAs upregulated alongside PRMT1 in patients with asthma in response to transforming growth factor beta 1 (TGF-β1).86 These findings suggest a role for PRMT1 in acute and chronic asthma in epithelial cells and fibroblasts. Furthermore, these findings have implications for the treatment of acute and chronic asthma as PRMT1 could serve as a specific therapeutic target.
Systemic Lupus Erythematosus
Elevated free aDMA levels are observed in the blood of patients with SLE.87 Further, elevated aDMA levels correlates with higher incidence of cardiovascular disease in these patients.86 Autoantibodies targeting the spliceosomal, RNA-binding Sm proteins are present in the serum of SLE patients.88 These autoantibodies recognize the symmetrically dimethylated RGG/RG epitopes of Sm proteins and p80-coilin,89 suggesting that inhibition of PRMTs may suppress some of their ability to induce autoimmune reactions.
Multiple Sclerosis
Citrullination of myelin basic protein (MBP) is a well-characterized post-translational modification required for myelin membrane stability. Increased citrullinated MBP is observed in patients with MS.90 The monomethylation and symmetric dimethylation of MBP at R107 was shown to be catalyzed by PRMT5.91,92 Early studies found that PRMT activity increases during the myelination phase of development and is a requirement for the formation of compact myelin in the CNS.93,94 Further, the importance of MBP methylation was demonstrated when myelinolysis was found to be associated with disturbances in methionine biosynthesis.95 Numerous animal and patient studies confirmed this observation and demonstrated that supplementation, particularly of vitamin B12, could reverse the degeneration. The PRMT5 inhibitor, HLCL65, was shown to effectively suppress adaptive memory Th cell responses and reduce inflammation in an EAE mouse model.96 PRMT5 depletion in CD4+ T cells was shown to protect mice from Th17-mediated diseases.73 These findings define a function for PRMT5 in Th cell expansion and its inhibition in inflammatory diseases caused by aberrant Th cell activity. A new study showed a correlation between EAE severity and PRMT5-mediated promotion of G₁/S cell cycle progression in CD4+ cells.97 These results corroborate the findings of Webb et al. 202073 and emphasize the importance PRMT5 inhibitors could have in suppressing Th cell expansion. The mechanism by which PRMT5 specifically promotes Th cell activity is not yet fully understood.
Acute Graft-Versus-Host Disease
The PRMT5 inhibitor, C220, can suppress T cell proliferation and cytokine production to alleviate the severity of aGVHD in mouse models having received hematopoietic cell transplants.98 PRMT5 inhibition deregulated the phosphorylation of extracellular signal-regulated kinase (ERK) 1 (ERK1), ERK2, and STAT1.98 Patients with lymphoma and acute myeloid leukemia (AML) who receive hematopoietic cell transplants may also benefit from the anti-tumoral activity of PRMT5 inhibitors.99,100
Ulcerative Colitis
The importance of arginine methylation in T lymphocytes is shown in ulcerative colitis.101 Mechanistically, PRMT5 depletion was found to indirectly lead to a decrease in H3K27 lysine methylation and DNMT1 binding at the Foxp3 promoter to support T regulatory (Treg) cell differentiation in ulcerative colitis patients as well as in clinical mouse models.101 PRMT5 mediates crosstalk with histone lysine methylation as H3R2me2s/H3R8me2s is needed for optimal deposition of methyl groups at H3K27 by enhancer of zeste homolog 2 (EZH2)/polycomb repressive complex 2 (PRC2).102 Thus, selective PRMT5 inhibition may be an effective therapeutic strategy to reduce intestinal inflammation.
Rheumatoid Arthritis
The PRMT5 inhibitor, EPZ015666, inhibits proliferation, migration, and invasion of fibroblast‐like synoviocytes (FLSs) from patients with RA by effectively reducing interleukin expression via the NF-κB and Ak strain transforming (AKT) pathways.41 These results demonstrate a unique role for PRMT5 in the context of RA and suggest that its specific inhibition may have therapeutic benefits for this autoimmune disease. Moreover, EPZ015666 attenuated cartilage degeneration in mouse models of osteoarthritis (OA).103 Furthermore, they show that PRMT5 overexpression in chondrocytes leads to elevated expression of matrix degrading enzymes via activation of MAPK and NF-κB signaling pathways. These data support the notion that PRMT5 inhibitors could have therapeutic value in the treatment of RA. Finally, post-translational modifications are known to play an important role in altering the immunogenicity of synovial tissue proteins. Namely, citrullination of type II collagen, α-enolase, and fibrinogen have been identified in patients with RA.104 Moreover, autoantibodies recognizing these citrullinated proteins have been found in the serum of patients with RA.105 Autoantibodies against methylated arginine epitopes have not yet been identified in patients with RA, however, it was reported that these patients have elevated levels of circulating sDMA metabolite in their circulation.106 These findings suggest that post-translational modifications may be a source of neoepitope production during inflammation.
PRMTs in Cancer Immunotherapy
PRMTs have recently been identified as regulators of cancer immunity. Here, we discuss the most recent advances involving arginine methylation in immune checkpoint pathways.
PRMT1
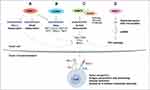
Deletion of PRMT1 using CRISPR-Cas9 sensitizes the colon adenocarcinoma cell line, MC38, to anti-programmed cell death protein-1 (PD-1) immunotherapy.107 Inhibiting PRMT1 was shown to sensitize tumors to T cell-mediated killing by enhancing the apoptosis of cancer cells. Furthermore, transcriptomic analysis showed that PRMT1 knockout alters the expression of genes involved in T cell-mediated tumor apoptosis and in the production of cytokines and chemokines such as CCL7 and CCL9.107 In the same context, the combination of a type I PRMT inhibitor, PT1001B, with anti-programmed death-ligand (PD-L) 1 (PD-L1) inhibition was shown to reduce pancreatic cancer progression by upregulating CD8+ T cell infiltration into tumors.108 PT1001B inhibits PD-L1 expression in cancer cells and enhances the induction of tumor cell apoptosis (Figure 4A).108 C57BL/6J mice injected with MC38 tumor cells and treated with type I PRMT inhibitor, MS023, also exhibit anti-tumor immunity. MS023 significantly inhibits tumor growth and enhances the checkpoint blockade.109 In human hepatocellular carcinoma (HCC), PRMT1 overexpression is associated with poor prognosis. Moreover, PRMT1 expression correlates with PD-L1 and PD-L2, suggesting that PRMT1 is an important regulator of immune checkpoint pathways in HCC.110
CARM1
A CRISPR-Cas9 screen identified that CARM1 deletion in the tumor enhances antitumoral immunity associated with an increase in CD8+ T cell and dendritic cell infiltration.63 CARM1 was identified as a negative regulator of tumor-specific T cells in the B16.F10 melanoma model. Another recent study has shown that inhibiting CARM1 with a chemical probe, compound 43 (a modified version of EZM2302),111 inhibited solid tumor growth of triple negative breast cancer cell line, BT549, and the melanoma cell line, A375, as xenografts in BALB/c nude mice. Compound 43 and EZM2302 displayed similar pharmacokinetic parameters, but compound 43 has a longer half-life and a higher plasma concentration. Zhang et al. 2021 showed that compound 43 exhibits excellent metabolic stability and elicits antitumor efficacy by increasing the number of activated CD8+ T cells, thereby regulating the immunosuppressive tumor microenvironment.112 These observations suggest that the inhibition of CARM1 may be used to treat solid tumors and be beneficial for the enhancement of cancer immunotherapy.
Recently, Fedoriw et al. 2022 showed that inhibiting type I PRMTs in cancer cells promotes antitumor immune responses by increasing T cell infiltration into the tumor microenvironment and enhancing the cytotoxic activity of T cells.113 Moreover, they showed that type I PRMT inhibitors increased the expression of interferon stimulated genes (ISGs) via the cGAS/stimulator of interferon genes (STING) cytosolic DNA sensing pathway (Figure 4A and B). They also showed reduced expression of the immunosuppressive factor, vascular endothelial growth factor (VEGF) (Figure 4A).113 Furthermore, combining type I PRMT inhibitors with immune checkpoint blockade enhanced the efficacy of cancer immunotherapy.113
PRMT5
PRMT5 has a pro-tumor intrinsic function as it promotes immunosuppression in melanoma mouse models. Notably, PRMT5 inhibition can potentiate immunotherapy by increasing IFN and chemokine production. It regulates the transcription of NOD-like receptor (NLR)-family caspase activation and recruitment domain (CARD)-containing 5 (NLRC5), a known regulator of the MHC-I antigen presentation pathway (Figure 4C).114 PRMT5 methylates forkhead box P3 (FOXP3), a transcription factor known to regulate Treg development and function.115 In this capacity, PRMT5 inhibition promotes tumor immunity by inhibiting Treg function and limiting Treg migration into tumors, thus, leading to enhancement of cancer immunotherapy and tumor-targeted therapies.
PRMT7
Recently, PRMT7 was identified as a new target to sensitize melanoma cells to cancer immunotherapy.116 It was shown that combining anti-PD-1 and anti-cytotoxic T lymphocyte-associated protein 4 (CTLA-4) therapy with PRMT7 deletion or PRMT7 inhibition using SGC3027, a cell permeable prodrug,117 enhances anti-tumor responsiveness to the immune checkpoint blockade in a melanoma mouse model.116 PRMT7-deficient B16.F10 melanoma cells exhibit increased dsRNA repetitive element expression, mimicking viral induction of the RIG-I pathway (Figure 4D). This induces type I IFNs and pro-inflammatory cytokines to enhance anti-tumoral immunity.116
In sum, the significance of PRMTs in the regulation of cancer immunity is gaining momentum and paving the way for future studies on the modulation and inhibition of arginine methylation for the treatment of solid tumors and to enhance the effectiveness of immune checkpoint blockade therapies.
PRMTs in Anti-Viral Responses
PRMTs are largely understood to negatively regulate the antiviral immune response.3 In this section, we will discuss recent advances involving PRMTs in this response.
PRMT1 was shown to directly interact with TANK-binding kinase-1 (TBK1) and catalyze its methylation to promote TBK1 phosphorylation and activation for IFN production.118 In contrast, type I PRMT inhibition in cancer cells stimulates IFN production,113 suggesting that PRMT1 regulation of the anti-viral response maybe context-dependent and cell-type-specific. Thus, myeloid-specific PRMT1 knockout mice are more susceptible to viral infections due to their inability to activate TBK1 signaling.118 Zebrafish PRMT3 and PRMT7 negatively regulate the antiviral response via IRF-3-mediated IFN production.119,120 Mammalian PRMT7 was shown to negatively regulate the antiviral response through the monomethylation of mitochondrial antiviral signaling protein (MAVS) and, thus, the RIG-I-like receptor (RLR) signaling pathway.121 PRMT5-mediated methylation of cGAS was also shown to abolish its DNA binding ability and attenuate the antiviral response via the cGAS/STING cytosolic DNA sensing pathway.114,122 PRMT5 additionally regulates the cGAS/STING pathway by methylating one of its components, the IFN-γ-inducible protein 16 (IFI16).114 Finally, it was shown that nuclear cGAS recruits PRMT5 and facilitates H3R2 methylation at the promoters of type I IFN genes, in turn enhancing antiviral immunity upon infection.123 Another study showed that PRMT5 activates the transcription of type I and type III IFNs, IFNβ1 and IFNλ1, via the induction of the activating transcription factor 2 (ATF2), c-Jun, and TBK1.71 Finally, PRMT6 was identified as a negative regulator of the TBK1-IRF3 signaling cascade attenuating the antiviral immune response.124,125
Arginine Methylation During Viral Infections
During viral infection both host and viral proteins are targets of PRMTs and their methylation has a profound influence on viral replication. PRMT6 was found to restrict human immunodeficiency virus (HIV) replication via the methylation of the HIV trans-activator (Tat) protein.126 Subsequent studies suggest that arginine methylation suppresses Tat-mediated transactivation by preventing its nucleolar retention and proteasomal degradation.127,128 Further, PRMT6 was found to methylate the HIV nucleocapsid protein and inhibit viral transcription.129 PRMT5 and PRMT7 were discovered to similarly support HIV replication via maintenance of viral protein R (Vpr) stability.130 Recently, inhibition of PRMT5 using EPZ015666 was shown to increase HIV internal ribosome entry site (IRES) activity via the loss of methylation of heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1).131 PRMT5 has been shown to methylate the C-terminal domain of hepatitis B virus (HBV) core (HBc) protein and repress viral replication and transcription.132 Certain viruses such as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and EBV may benefit from PRMT inhibition. Briefly, the SARS-CoV-2 nucleocapsid (N) protein was recently shown to be methylated by PRMT1.133 Interestingly, PRMT1 depletion or inhibition using MS023 was found to interfere with the ability of the N protein to localize to stress granules and bind to the 5’-UTR of SARS-CoV-2 RNA.133 Furthermore, MS023 was shown to reduce SARS-CoV-2 replication in VeroE6 cells.133 Finally, PRMT5 activity is understood to play a crucial role in Epstein-Barr nuclear antigen (EBNA) 1 (EBNA1)-mediated viral replication, EBNA2-mediated transcription, and EBV-dependent B cell immortalization.134 Thus, PRMT5 inhibition might prove to be an effective therapeutic strategy in the treatment of EBV.
Although accumulating evidence implicates PRMTs in the modulation of anti-viral immunity, the underlying mechanisms by which arginine methylation regulates the antiviral immune response is not fully understood. Continued investigation will reveal these mechanisms and aid in developing more effective anti-viral treatments and diagnostic tools.
PRMT Inhibitors in the Treatment of Immune and Inflammatory-Related Diseases
There are several small molecule PRMT inhibitors currently in clinical trials for the treatment of cancer (for review see3). This is not surprising considering the numerous roles that PRMTs play in the regulation of cancer, the immune system, and anti-viral and inflammatory responses. PRMT inhibitors are therefore attractive therapeutic targets, particularly in lymphomas and leukemias where their expression is elevated.
A first-in-class PRMT5 degrader, MS4322, using PROTAC, has been developed.47 MS4322 effectively degrades PRMT5 in an E3 ligase- and proteasome-dependent manner in mammalian cells.47 Another approach has been to block the PRMT5-substrate adaptor interaction with first-in-class PRMT5 binding motif (PBM)-competitive small molecule, BRD0639. This inhibitor was shown to effectively outcompete binding between PRMT5 and RIOK1, inhibiting the methylation of certain PRMT5 substrates dependent on RIOK1 interaction.135 A new potent and selective PRMT5 inhibitor that binds to the MTA-bound PRMT5 complex has also been developed. MRTX1719 has been shown to inhibit PRMT5 activity exclusively in MTAP negative cells.46 This compound allows for selective targeting of only MTA-bound PRMT5 in MTAP negative cancer cells.
Several PRMT inhibitors target more than one PRMT.136 For example, MS023 and GSK3368715 are both general type I PRMT inhibitors.99,137 While MS049 targets both CARM1 and PRMT6, and DS-437 is a dual PRMT5 and PRMT7 inhibitor.43,138 Finally, EPZ020411 has a higher affinity for PRMT6, but can also inhibit PRMT1 and PRMT8.139 We summarize the main features of small-molecule PRMT inhibitors discussed herein as well as those that have been investigated in the context of immune and inflammatory-related diseases in Table 1.
Conclusions and Future Perspectives
As discussed in this review, arginine methylation plays an essential role in regulating inflammation, immunity, and antiviral responses, in particular, by modulating the activity of NF-κB. Although more detailed analyses are required, multiple studies propose that PRMTs can be targeted to improve inflammatory-related diseases, as well as leukemias and lymphomas. The development of drugs targeting the activity of PRMTs has gained significant momentum in the last several years, and the inclusion of PRMT inhibitors in current clinical trials warrants continued research on arginine methylation. The prospect of using PRMT inhibitors as anti-inflammatory and/or anti-viral, besides their use as potential cancer therapeutics, is promising.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Blanc RS, Richard S. Arginine methylation: the coming of age. Mol Cell. 2017;65(1):8–24. doi:10.1016/j.molcel.2016.11.003
2. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33(1):1–13. doi:10.1016/j.molcel.2008.12.013
3. Xu J, Richard S. Cellular pathways influenced by protein arginine methylation: implications for cancer. Mol Cell. 2021;81(21):4357–4368. doi:10.1016/j.molcel.2021.09.011
4. Jambhekar A, Anastas JN, Shi Y. Histone lysine demethylase inhibitors. Cold Spring Harb Perspect Med. 2017;7(1):a026484. doi:10.1101/cshperspect.a026484
5. Webby CJ, Wolf A, Gromak N, et al. Jmjd6 catalyses lysyl-hydroxylation of U2AF65, a protein associated with RNA splicing. Science. 2009;325(5936):90–93. doi:10.1126/science.1175865
6. Walport LJ, Hopkinson RJ, Chowdhury R, et al. Arginine demethylation is catalysed by a subset of JmjC histone lysine demethylases. Nat Commun. 2016;7:11974. doi:10.1038/ncomms11974
7. Mondal S, Thompson PR. Chemical biology of protein citrullination by the protein A arginine deiminases. Curr Opin Chem Biol. 2021;63:19–27. doi:10.1016/j.cbpa.2021.01.010
8. Gayatri S, Bedford MT. Readers of histone methylarginine marks. Biochim Biophys Acta. 2014;1839(8):702–710. doi:10.1016/j.bbagrm.2014.02.015
9. Guccione E, Richard S. The regulation, functions and clinical relevance of arginine methylation. Nat Rev Mol Cell Biol. 2019;20(10):642–657. doi:10.1038/s41580-019-0155-x
10. Zhang X, Bai XC, Chen ZJ. Structures and mechanisms in the cGAS-STING innate immunity pathway. Immunity. 2020;53(1):43–53. doi:10.1016/j.immuni.2020.05.013
11. Liu T, Zhang L, Joo D, Sun SC. NF-kappaB signaling in inflammation. Signal Transduct Target Ther. 2017;2. doi:10.1038/sigtrans.2017.23
12. Bjorkstrom NK, Strunz B, Ljunggren HG. Natural killer cells in antiviral immunity. Nat Rev Immunol. 2021;22:112–123. doi:10.1038/s41577-021-00558-3
13. Ghasemi N, Razavi S, Nikzad E. Multiple sclerosis: pathogenesis, symptoms, diagnoses and cell-based therapy. Cell J. 2017;19(1):1–10. doi:10.22074/cellj.2016.4867
14. Guo Q, Wang Y, Xu D, Nossent J, Pavlos NJ, Xu J. Rheumatoid arthritis: pathological mechanisms and modern pharmacologic therapies. Bone Res. 2018;6:15. doi:10.1038/s41413-018-0016-9
15. Kudo M, Ishigatsubo Y, Aoki I. Pathology of asthma. Front Microbiol. 2013;4:263. doi:10.3389/fmicb.2013.00263
16. Pawlak MR, Scherer CA, Chen J, Roshon MJ, Ruley HE. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol Cell Biol. 2000;20(13):4859–4869. doi:10.1128/mcb.20.13.4859-4869.2000
17. Wang H, Huang ZQ, Xia L, et al. Methylation of histone H4 at arginine 3 facilitating transcriptional activation by nuclear hormone receptor. Science. 2001;293(5531):853–857. doi:10.1126/science.1060781
18. Kim JH, Yoo BC, Yang WS, Kim E, Hong S, Cho JY. The role of protein arginine methyltransferases in inflammatory responses. Mediators Inflamm. 2016;2016:4028353. doi:10.1155/2016/4028353
19. Fan Z, Li J, Li P, et al. Protein arginine methyltransferase 1 (PRMT1) represses MHC II transcription in macrophages by methylating CIITA. Sci Rep. 2017;7:40531. doi:10.1038/srep40531
20. Reintjes A, Fuchs JE, Kremser L, et al. Asymmetric arginine dimethylation of RelA provides a repressive mark to modulate TNFalpha/NF-kappaB response. Proc Natl Acad Sci U S A. 2016;113(16):4326–4331. doi:10.1073/pnas.1522372113
21. Kleinschmidt MA, Streubel G, Samans B, Krause M, Bauer UM. The protein arginine methyltransferases CARM1 and PRMT1 cooperate in gene regulation. Nucleic Acids Res. 2008;36(10):3202–3213. doi:10.1093/nar/gkn166
22. Lou X, Sun S, Chen W, et al. Negative feedback regulation of NF-kappaB action by CITED2 in the nucleus. J Immunol. 2011;186(1):539–548. doi:10.4049/jimmunol.1001650
23. Kryukov GV, Wilson FH, Ruth JR, et al. MTAP deletion confers enhanced dependency on the PRMT5 arginine methyltransferase in cancer cells. Science. 2016;351(6278):1214–1218. doi:10.1126/science.aad5214
24. Mavrakis KJ, McDonald ER, Schlabach MR, et al. Disordered methionine metabolism in MTAP/CDKN2A-deleted cancers leads to dependence on PRMT5. Science. 2016;351(6278):1208–1213. doi:10.1126/science.aad5944
25. Marjon K, Cameron MJ, Quang P, et al. MTAP deletions in cancer create vulnerability to targeting of the MAT2A/PRMT5/RIOK1 axis. Cell Rep. 2016;15(3):574–587. doi:10.1016/j.celrep.2016.03.043
26. Browne SK, Roesser JR, Zhu SZ, Ginder GD. Differential IFN-gamma stimulation of HLA-A gene expression through CRM-1-dependent nuclear RNA export. J Immunol. 2006;177(12):8612–8619. doi:10.4049/jimmunol.177.12.8612
27. Lafleur VN, Richard S, Richard DE. Transcriptional repression of hypoxia-inducible factor-1 (HIF-1) by the protein arginine methyltransferase PRMT1. Mol Biol Cell. 2014;25(6):925–935. doi:10.1091/mbc.E13-07-0423
28. Gupta P, Singh A, Gowda P, Ghosh S, Chatterjee A, Sen E. Lactate induced HIF-1alpha-PRMT1 cross talk affects MHC I expression in monocytes. Exp Cell Res. 2016;347(2):293–300. doi:10.1016/j.yexcr.2016.08.008
29. Mowen KA, Schurter BT, Fathman JW, David M, Glimcher LH. Arginine methylation of NIP45 modulates cytokine gene expression in effector T lymphocytes. Mol Cell. 2004;15(4):559–571. doi:10.1016/j.molcel.2004.06.042
30. Fathman JW, Gurish MF, Hemmers S, et al. NIP45 controls the magnitude of the type 2 T helper cell response. Proc Natl Acad Sci U S A. 2010;107(8):3663–3668. doi:10.1073/pnas.0914700107
31. Koch S, Knipfer L, Kolle J, et al. Targeted deletion of NFAT-interacting-protein-(NIP) 45 resolves experimental asthma by inhibiting innate lymphoid cells group 2 (ILC2). Sci Rep. 2019;9(1):15695. doi:10.1038/s41598-019-51690-z
32. Stopa N, Krebs JE, Shechter D. The PRMT5 arginine methyltransferase: many roles in development, cancer and beyond. Cell Mol Life Sci. 2015;72(11):2041–2059. doi:10.1007/s00018-015-1847-9
33. Pollack BP, Kotenko SV, He W, Izotova LS, Barnoski BL, Pestka S. The human homologue of the yeast proteins Skb1 and Hsl7p interacts with Jak kinases and contains protein methyltransferase activity. J Biol Chem. 1999;274(44):31531–31542. doi:10.1074/jbc.274.44.31531
34. Friesen WJ, Paushkin S, Wyce A, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21(24):8289–8300. doi:10.1128/MCB.21.24.8289-8300.2001
35. Tanaka H, Hoshikawa Y, Oh-hara T, et al. PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-kappaB activation. Mol Cancer Res. 2009;7(4):557–569. doi:10.1158/1541-7786.Mcr-08-0197
36. Tanaka Y, Nagai Y, Okumura M, Greene MI, Kambayashi T. PRMT5 is required for T cell survival and proliferation by maintaining cytokine signaling. Front Immunol. 2020;11:621. doi:10.3389/fimmu.2020.00621
37. Bandyopadhyay S, Harris DP, Adams GN, et al. HOXA9 methylation by PRMT5 is essential for endothelial cell expression of leukocyte adhesion molecules. Mol Cell Biol. 2012;32(7):1202–1213. doi:10.1128/mcb.05977-11
38. Harris DP, Bandyopadhyay S, Maxwell TJ, Willard B, DiCorleto PE. Tumor necrosis factor (TNF)-alpha induction of CXCL10 in endothelial cells requires protein arginine methyltransferase 5 (PRMT5)-mediated nuclear factor (NF)-kappaB p65 methylation. J Biol Chem. 2014;289(22):15328–15339. doi:10.1074/jbc.M114.547349
39. Harris DP, Chandrasekharan UM, Bandyopadhyay S, Willard B, DiCorleto PE. PRMT5-mediated methylation of NF-kappaB p65 at Arg174 is required for endothelial CXCL11 gene induction in response to TNF-alpha and IFN-gamma costimulation. PLoS One. 2016;11(2):e0148905. doi:10.1371/journal.pone.0148905
40. Wei H, Wang B, Miyagi M, et al. PRMT5 dimethylates R30 of the p65 subunit to activate NF-kappaB. Proc Natl Acad Sci U S A. 2013;110(33):13516–13521. doi:10.1073/pnas.1311784110
41. Chen D, Zeng S, Huang M, Xu H, Liang L, Yang X. Role of protein arginine methyltransferase 5 in inflammation and migration of fibroblast-like synoviocytes in rheumatoid arthritis. J Cell Mol Med. 2017;21(4):781–790. doi:10.1111/jcmm.13020
42. Liu H, Jia K, Ren Z, Sun J, Pan LL. PRMT5 critically mediates TMAO-induced inflammatory response in vascular smooth muscle cells. Cell Death Dis. 2022;13(4):299. doi:10.1038/s41419-022-04719-7
43. Smil D, Eram MS, Li F, et al. Discovery of a dual PRMT5-PRMT7 inhibitor. ACS Med Chem Lett. 2015;6(4):408–412. doi:10.1021/ml500467h
44. Chan-Penebre E, Kuplast KG, Majer CR, et al. A selective inhibitor of PRMT5 with in vivo and in vitro potency in MCL models. Nat Chem Biol. 2015;11(6):432–437. doi:10.1038/nchembio.1810
45. Gerhart SV, Kellner WA, Thompson C, et al. Activation of the p53-MDM4 regulatory axis defines the anti-tumour response to PRMT5 inhibition through its role in regulating cellular splicing. Sci Rep. 2018;8(1):9711. doi:10.1038/s41598-018-28002-y
46. Smith CR, Aranda R, Bobinski TP, et al. Fragment-based discovery of MRTX1719, a synthetic lethal inhibitor of the PRMT5•MTA complex for the treatment of MTAP -deleted cancers. J Med Chem. 2022;65:1749–1766. doi:10.1021/acs.jmedchem.1c01900
47. Shen Y, Gao G, Yu X, et al. Discovery of first-in-class protein arginine methyltransferase 5 (PRMT5) degraders. J Med Chem. 2020;63(17):9977–9989. doi:10.1021/acs.jmedchem.0c01111
48. Mulvaney KM, Blomquist C, Acharya N, et al. Molecular basis for substrate recruitment to the PRMT5 methylosome. Mol Cell. 2021;81(17):3481–3495e7. doi:10.1016/j.molcel.2021.07.019
49. Di Lorenzo A, Yang Y, Macaluso M, Bedford MT. A gain-of-function mouse model identifies PRMT6 as a NF-kappaB coactivator. Nucleic Acids Res. 2014;42(13):8297–8309. doi:10.1093/nar/gku530
50. Covic M, Hassa PO, Saccani S, et al. Arginine methyltransferase CARM1 is a promoter-specific regulator of NF-kappaB-dependent gene expression. EMBO J. 2005;24(1):85–96. doi:10.1038/sj.emboj.7600500
51. Miao F, Li S, Chavez V, Lanting L, Natarajan R. Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol Endocrinol. 2006;20(7):1562–1573. doi:10.1210/me.2005-0365
52. Greenblatt SM, Liu F, Nimer SD. Arginine methyltransferases in normal and malignant hematopoiesis. Exp Hematol. 2016;44(6):435–441. doi:10.1016/j.exphem.2016.03.009
53. Sen S, He Z, Ghosh S, et al. PRMT1 plays a critical role in Th17 differentiation by regulating reciprocal recruitment of STAT3 and STAT5. J Immunol. 2018;201(2):440–450. doi:10.4049/jimmunol.1701654
54. Vadnais C, Chen R, Fraszczak J, et al. GFI1 facilitates efficient DNA repair by regulating PRMT1 dependent methylation of MRE11 and 53BP1. Nat Commun. 2018;9(1):1418. doi:10.1038/s41467-018-03817-5
55. Bonham K, Hemmers S, Lim YH, Hill DM, Finn MG, Mowen KA. Effects of a novel arginine methyltransferase inhibitor on T-helper cell cytokine production. FEBS J. 2010;277(9):2096–2108. doi:10.1111/j.1742-4658.2010.07623.x
56. Dolezal E, Infantino S, Drepper F, et al. The BTG2-PRMT1 module limits pre-B cell expansion by regulating the CDK4-Cyclin-D3 complex. Nat Immunol. 2017;18(8):911–920. doi:10.1038/ni.3774
57. Hata K, Yanase N, Sudo K, et al. Differential regulation of T-cell dependent and T-cell independent antibody responses through arginine methyltransferase PRMT1 in vivo. FEBS Lett. 2016;590(8):1200–1210. doi:10.1002/1873-3468.12161
58. Infantino S, Benz B, Waldmann T, Jung M, Schneider R, Reth M. Arginine methylation of the B cell antigen receptor promotes differentiation. J Exp Med. 2010;207(4):711–719. doi:10.1084/jem.20091303
59. Infantino S, Light A, O’Donnell K, et al. Arginine methylation catalyzed by PRMT1 is required for B cell activation and differentiation. Nat Commun. 2017;8(1):891. doi:10.1038/s41467-017-01009-1
60. Zhao J, O’Neil M, Vittal A, Weinman SA, Tikhanovich I. PRMT1-dependent macrophage IL-6 production is required for alcohol-induced HCC progression. Gene Expr. 2019;19(2):137–150. doi:10.3727/105221618x15372014086197
61. Kim J, Lee J, Yadav N, et al. Loss of CARM1 results in hypomethylation of thymocyte cyclic AMP-regulated phosphoprotein and deregulated early T cell development. J Biol Chem. 2004;279(24):25339–25344. doi:10.1074/jbc.M402544200
62. Kim D, Lee J, Cheng D, et al. Enzymatic activity is required for the in vivo functions of CARM1. J Biol Chem. 2010;285(2):1147–1152. doi:10.1074/jbc.M109.035865
63. Kumar S, Zeng Z, Bagati A, et al. CARM1 inhibition enables immunotherapy of resistant tumors by dual action on tumor cells and T cells. Cancer Discov. 2021;11(8):2050–2071. doi:10.1158/2159-8290.Cd-20-1144
64. Raghu D, Xue HH, Mielke LA. Control of lymphocyte fate, infection, and tumor immunity by TCF-1. Trends Immunol. 2019;40(12):1149–1162. doi:10.1016/j.it.2019.10.006
65. Gautam S, Fioravanti J, Zhu W, et al. The transcription factor c-Myb regulates CD8(+) T cell stemness and antitumor immunity. Nat Immunol. 2019;20(3):337–349. doi:10.1038/s41590-018-0311-z
66. Sen S, Wang F, Zhang J, et al. SRC1 promotes Th17 differentiation by overriding Foxp3 suppression to stimulate RORgammat activity in a PKC-theta-dependent manner. Proc Natl Acad Sci U S A. 2018;115(3):E458–E467. doi:10.1073/pnas.1717789115
67. Lai Y, Li X, Li T, et al. Protein arginine N-methyltransferase 4 (PRMT4) contributes to lymphopenia in experimental sepsis. Thorax. 2022;
68. Vu LP, Perna F, Wang L, et al. PRMT4 blocks myeloid differentiation by assembling a methyl-RUNX1-dependent repressor complex. Cell Rep. 2013;5(6):1625–1638. doi:10.1016/j.celrep.2013.11.025
69. Greenblatt SM, Man N, Hamard PJ, et al. CARM1 is essential for myeloid leukemogenesis but dispensable for normal hematopoiesis. Cancer Cell. 2018;33(6):1111–1127e5. doi:10.1016/j.ccell.2018.05.007
70. Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem J. 2005;388(Pt1):379–386. doi:10.1042/bj20040373
71. Metz PJ, Ching KA, Xie T, et al. Symmetric arginine dimethylation is selectively required for mRNA splicing and the initiation of type I and type III interferon signaling. Cell Rep. 2020;30(6):1935–1950 e8. doi:10.1016/j.celrep.2020.01.054
72. Inoue M, Okamoto K, Terashima A, et al. Arginine methylation controls the strength of γc-family cytokine signaling in T cell maintenance. Nat Immunol. 2018;19(11):1265–1276. doi:10.1038/s41590-018-0222-z
73. Webb LM, Sengupta S, Edell C, et al. Protein arginine methyltransferase 5 promotes cholesterol biosynthesis-mediated Th17 responses and autoimmunity. J Clin Invest. 2020;130(4):1683–1698. doi:10.1172/JCI131254
74. Panfil AR, Al-Saleem J, Howard CM, et al. PRMT5 is upregulated in HTLV-1-mediated T-cell transformation and selective inhibition alters viral gene expression and infected cell survival. Viruses. 2015;8(1):7. doi:10.3390/v8010007
75. Igarashi H, Kuwahara K, Yoshida M, et al. GANP suppresses the arginine methyltransferase PRMT5 regulating IL-4-mediated STAT6-signaling to IgE production in B cells. Mol Immunol. 2009;46(6):1031–1041. doi:10.1016/j.molimm.2008.08.272
76. Alinari L, Mahasenan KV, Yan F, et al. Selective inhibition of protein arginine methyltransferase 5 blocks initiation and maintenance of B-cell transformation PRMT5 promotes Cyclin E1 and cell cycle progression in CD4 Th1 cells and correlates with EAE severity. Blood. 2015;125(16):2530–2543. doi:10.1182/blood-2014-12-619783
77. Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24(21):9630–9645. doi:10.1128/MCB.24.21.9630-9645.2004
78. Majumder S, Alinari L, Roy S, et al. Methylation of histone H3 and H4 by PRMT5 regulates ribosomal RNA gene transcription. J Cell Biochem. 2010;109(3):553–563. doi:10.1002/jcb.22432
79. Liu F, Cheng G, Hamard PJ, et al. Arginine methyltransferase PRMT5 is essential for sustaining normal adult hematopoiesis. J Clin Invest. 2015;125(9):3532–3544. doi:10.1172/jci81749
80. Litzler LC, Zahn A, Meli AP, et al. PRMT5 is essential for B cell development and germinal center dynamics. Nat Commun. 2019;10(1):22. doi:10.1038/s41467-018-07884-6
81. Ying Z, Mei M, Zhang P, et al. Histone arginine methylation by PRMT7 controls germinal center formation via regulating Bcl6 transcription. J Immunol. 2015;195(4):1538–1547. doi:10.4049/jimmunol.1500224
82. Jain K, Jin CY, Clarke SG. Epigenetic control via allosteric regulation of mammalian protein arginine methyltransferases. Proc Natl Acad Sci U S A. 2017;114(38):10101–10106. doi:10.1073/pnas.1706978114
83. Sun Q, Liu L, Roth M, et al. PRMT1 upregulated by epithelial proinflammatory cytokines participates in COX2 expression in fibroblasts and chronic antigen-induced pulmonary inflammation. J Immunol. 2015;195(1):298–306. doi:10.4049/jimmunol.1402465
84. Sun Q, Yang X, Zhong B, et al. Upregulated protein arginine methyltransferase 1 by IL-4 increases eotaxin-1 expression in airway epithelial cells and participates in antigen-induced pulmonary inflammation in rats. J Immunol. 2012;188(7):3506–3512. doi:10.4049/jimmunol.1102635
85. Liu L, Sun Q, Bao R, et al. Specific regulation of PRMT1 expression by PIAS1 and RKIP in BEAS-2B epithelia cells and HFL-1 fibroblasts in lung inflammation. Sci Rep. 2016;6:21810. doi:10.1038/srep21810
86. Zhai W, Sun H, Li Z, et al. PRMT1 modulates processing of asthma-related primary MicroRNAs (Pri-miRNAs) into mature miRNAs in lung epithelial cells. J Immunol. 2021;206(1):11–22. doi:10.4049/jimmunol.2000887
87. Bultink IE, Teerlink T, Heijst JA, Dijkmans BA, Voskuyl AE. Raised plasma levels of asymmetric dimethylarginine are associated with cardiovascular events, disease activity, and organ damage in patients with systemic lupus erythematosus. Ann Rheum Dis. 2005;64(9):1362–1365. doi:10.1136/ard.2005.036137
88. Arbuckle MR, McClain MT, Rubertone MV, et al. Development of autoantibodies before the clinical onset of systemic lupus erythematosus. N Engl J Med. 2003;349(16):1526–1533. doi:10.1056/NEJMoa021933
89. Chang HH, Hu HH, Lee YJ, et al. Proteomic analyses and identification of arginine methylated proteins differentially recognized by autosera from anti-Sm positive SLE patients. J Biomed Sci. 2013;20:27. doi:10.1186/1423-0127-20-27
90. Beniac DR, Wood DD, Palaniyar N, Ottensmeyer FP, Moscarello MA, Harauz G. Cryoelectron microscopy of protein-lipid complexes of human myelin basic protein charge isomers differing in degree of citrullination. J Struct Biol. 2000;129(1):80–95. doi:10.1006/jsbi.1999.4200
91. Baldwin GS, Carnegie PR. Specific enzymic methylation of an arginine in the experimental allergic encephalomyelitis protein from human myelin. Science. 1971;171(3971):579–581. doi:10.1126/science.171.3971.579
92. Rho J, Choi S, Seong YR, Cho WK, Kim SH, Im DS. Prmt5, which forms distinct homo-oligomers, is a member of the protein-arginine methyltransferase family. J Biol Chem. 2001;276(14):11393–11401. doi:10.1074/jbc.M008660200
93. Amur SG, Shanker G, Cochran JM, Ved HS, Pieringer RA. Correlation between inhibition of myelin basic protein (arginine) methyltransferase by sinefungin and lack of compact myelin formation in cultures of cerebral cells from embryonic mice. J Neurosci Res. 1986;16(2):367–376. doi:10.1002/jnr.490160204
94. Crang AJ, Jacobson W. The relationship of myelin basic protein (arginine) methyltransferase to myelination in mouse spinal cord. J Neurochem. 1982;39(1):244–247. doi:10.1111/j.1471-4159.1982.tb04726.x
95. Kim S, Lim IK, Park GH, Paik WK. Biological methylation of myelin basic protein: enzymology and biological significance. Int J Biochem Cell Biol. 1997;29(5):743–751. doi:10.1016/s1357-2725(97)00009-5
96. Webb LM, Amici SA, Jablonski KA, et al. PRMT5-selective inhibitors suppress inflammatory T cell responses and experimental autoimmune encephalomyelitis. J Immunol. 2017;198(4):1439–1451. doi:10.4049/jimmunol.1601702
97. Amici SA, Osman W, Guerau-de-Arellano M. PRMT5 promotes cyclin E1 and cell cycle progression in CD4 Th1 cells and correlates with EAE severity. Front Immunol. 2021;12:695947. doi:10.3389/fimmu.2021.695947
98. Snyder KJ, Zitzer NC, Gao Y, et al. PRMT5 regulates T cell interferon response and is a target for acute graft-versus-host disease. JCI Insight. 2020;5(8). doi:10.1172/jci.insight.131099
99. Fedoriw A, Rajapurkar SR, O’Brien S, et al. Anti-tumor activity of the type I PRMT inhibitor, GSK3368715, synergizes with PRMT5 inhibition through MTAP loss. Cancer Cell. 2019;36(1):100–114e25. doi:10.1016/j.ccell.2019.05.014
100. Fong JY, Pignata L, Goy PA, et al. Therapeutic targeting of RNA splicing catalysis through inhibition of protein arginine methylation. Cancer Cell. 2019;36(2):194–209 e9. doi:10.1016/j.ccell.2019.07.003
101. Zheng Y, Huang L, Ge W, et al. Protein arginine methyltransferase 5 inhibition upregulates Foxp3(+) regulatory T cells frequency and function during the ulcerative colitis. Front Immunol. 2017;8:596. doi:10.3389/fimmu.2017.00596
102. Liu F, Xu Y, Lu X, et al. PRMT5-mediated histone arginine methylation antagonizes transcriptional repression by polycomb complex PRC2. Nucleic Acids Res. 2020;48(6):2956–2968. doi:10.1093/nar/gkaa065
103. Dong Y, Wang P, Yang Y, et al. PRMT5 inhibition attenuates cartilage degradation by reducing MAPK and NF-kappaB signaling. Arthritis Res Ther. 2020;22(1):201. doi:10.1186/s13075-020-02304-x
104. Burska AN, Hunt L, Boissinot M, et al. Autoantibodies to posttranslational modifications in rheumatoid arthritis. Mediators Inflamm. 2014;2014:492873. doi:10.1155/2014/492873
105. Snir O, Widhe M, von Spee C, et al. Multiple antibody reactivities to citrullinated antigens in sera from patients with rheumatoid arthritis: association with HLA-DRB1 alleles. Ann Rheum Dis. 2009;68(5):736–743. doi:10.1136/ard.2008.091355
106. Chandrasekharan UM, Wang Z, Wu Y, et al. Elevated levels of plasma symmetric dimethylarginine and increased arginase activity as potential indicators of cardiovascular comorbidity in rheumatoid arthritis. Arthritis Res Ther. 2018;20(1):123. doi:10.1186/s13075-018-1616-x
107. Hou J, Wang Y, Shi L, et al. Integrating genome-wide CRISPR immune screen with multi-omic clinical data reveals distinct classes of tumor intrinsic immune regulators. J Immunother Cancer. 2021;9(2). doi:10.1136/jitc-2020-001819
108. Zheng NN, Zhou M, Sun F, et al. Combining protein arginine methyltransferase inhibitor and anti-programmed death-ligand-1 inhibits pancreatic cancer progression. World J Gastroenterol. 2020;26(26):3737–3749. doi:10.3748/wjg.v26.i26.3737
109. Lu SX, De Neef E, Thomas JD, et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell. 2021;184(15):4032–4047 e31. doi:10.1016/j.cell.2021.05.038
110. Schonfeld M, Zhao J, Komatz A, Weinman SA, Tikhanovich I. The polymorphism rs975484 in the protein arginine methyltransferase 1 gene modulates expression of immune checkpoint genes in hepatocellular carcinoma. J Biol Chem. 2020;295(20):7126–7137. doi:10.1074/jbc.RA120.013401
111. Drew AE, Moradei O, Jacques SL, et al. Identification of a CARM1 inhibitor with potent in vitro and in vivo activity in preclinical models of multiple myeloma. Sci Rep. 2017;7(1):17993. doi:10.1038/s41598-017-18446-z
112. Zhang Z, Guo Z, Xu X, et al. Structure-based discovery of potent CARM1 inhibitors for solid tumor and cancer immunology therapy. J Med Chem. 2021;64(22):16650–16674. doi:10.1021/acs.jmedchem.1c01308
113. Fedoriw A, Shi L, O’Brien S, et al. Inhibiting type I arginine methyltransferase activity promotes the T cell mediated antitumor immune response. Cancer Immunol Res. 2022;10:420–436. doi:10.1158/2326-6066.CIR-21-0614
114. Kim H, Kim H, Feng Y, et al. PRMT5 control of cGAS/STING and NLRC5 pathways defines melanoma response to antitumor immunity. Sci Transl Med. 2020;12(551). doi:10.1126/scitranslmed.aaz5683
115. Nagai Y, Ji MQ, Zhu F, et al. PRMT5 associates with the FOXP3 homomer and when disabled enhances targeted p185(erbB2/neu) tumor immunotherapy. Front Immunol. 2019;10:174. doi:10.3389/fimmu.2019.00174
116. Srour N, Villarreal OD, Hardikar S, et al. PRMT7 ablation stimulates anti-tumor immunity and sensitizes melanoma to immune checkpoint blockade. Cell Rep. 2022;38(13):110582. doi:10.1016/j.celrep.2022.110582
117. Szewczyk MM, Ishikawa Y, Organ S, et al. Pharmacological inhibition of PRMT7 links arginine monomethylation to the cellular stress response. Nat Commun. 2020;11(1):2396. doi:10.1038/s41467-020-16271-z
118. Yan Z, Wu H, Liu H, et al. The protein arginine methyltransferase PRMT1 promotes TBK1 activation through asymmetric arginine methylation. Cell Rep. 2021;36(12):109731. doi:10.1016/j.celrep.2021.109731
119. Zhu J, Liu X, Cai X, et al. Zebrafish prmt7 negatively regulates antiviral responses by suppressing the retinoic acid-inducible gene-I-like receptor signaling. FASEB J. 2020;34(1):988–1000. doi:10.1096/fj.201902219R
120. Zhu J, Liu X, Cai X, et al. Zebrafish prmt3 negatively regulates antiviral responses. FASEB J. 2020;34(8):10212–10227. doi:10.1096/fj.201902569R
121. Zhu J, Li X, Cai X, et al. Arginine monomethylation by PRMT7 controls MAVS-mediated antiviral innate immunity. Mol Cell. 2021;81(15):3171–3186e8. doi:10.1016/j.molcel.2021.06.004
122. Ma D, Yang M, Wang Q, et al. Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Sci Adv. 2021;7(13). doi:10.1126/sciadv.abc1834
123. Cui S, Yu Q, Chu L, et al. Nuclear cGAS functions non-canonically to enhance antiviral immunity via recruiting methyltransferase Prmt5. Cell Rep. 2020;33(10):108490. doi:10.1016/j.celrep.2020.108490
124. Zhang H, Han C, Li T, Li N, Cao X. The methyltransferase PRMT6 attenuates antiviral innate immunity by blocking TBK1-IRF3 signaling. Cell Mol Immunol. 2019;16(10):800–809. doi:10.1038/s41423-018-0057-4
125. Jiang Y, Liu L, Yang S, et al. Black carp PRMT6 inhibits TBK1-IRF3/7 signaling during the antiviral innate immune activation. Fish Shellfish Immunol. 2019;93:108–115. doi:10.1016/j.fsi.2019.07.044
126. Boulanger MC, Liang C, Russell RS, et al. Methylation of Tat by PRMT6 regulates human immunodeficiency virus type 1 gene expression. J Virol. 2005;79(1):124–131. doi:10.1128/jvi.79.1.124-131.2005
127. Fulcher AJ, Sivakumaran H, Jin H, Rawle DJ, Harrich D, Jans DA. The protein arginine methyltransferase PRMT6 inhibits HIV-1 Tat nucleolar retention. Biochim Biophys Acta. 2016;1863(2):254–262. doi:10.1016/j.bbamcr.2015.11.019
128. Sivakumaran H, van der Horst A, Fulcher AJ, et al. Arginine methylation increases the stability of human immunodeficiency virus type 1 Tat. J Virol. 2009;83(22):11694–11703. doi:10.1128/jvi.00499-09
129. Invernizzi CF, Xie B, Frankel FA, et al. Arginine methylation of the HIV-1 nucleocapsid protein results in its diminished function. AIDS. 2007;21(7):795–805. doi:10.1097/QAD.0b013e32803277ae
130. Murakami H, Suzuki T, Tsuchiya K, et al. Protein arginine N-methyltransferases 5 and 7 promote HIV-1 production. Viruses. 2020;12(3):355. doi:10.3390/v12030355
131. Barrera A, Ramos H, Vera-Otarola J, et al. Post-translational modifications of hnRNP A1 differentially modulate retroviral IRES-mediated translation initiation. Nucleic Acids Res. 2020;48(18):10479–10499. doi:10.1093/nar/gkaa765
132. Lubyova B, Hodek J, Zabransky A, et al. PRMT5: a novel regulator of Hepatitis B virus replication and an arginine methylase of HBV core. PLoS One. 2017;12(10):e0186982. doi:10.1371/journal.pone.0186982
133. Cai T, Yu Z, Wang Z, Liang C, Richard S. Arginine methylation of SARS-Cov-2 nucleocapsid protein regulates RNA binding, its ability to suppress stress granule formation, and viral replication. J Biol Chem. 2021;297(1):100821. doi:10.1016/j.jbc.2021.100821
134. Liang Z, Wen C, Jiang H, Ma S, Liu X. Protein arginine methyltransferase 5 functions via interacting proteins. Front Cell Dev Biol. 2021;9:725301. doi:10.3389/fcell.2021.725301
135. McKinney DC, McMillan BJ, Ranaghan MJ, et al. Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction. J Med Chem. 2021;64(15):11148–11168. doi:10.1021/acs.jmedchem.1c00507
136. Hwang JW, Cho Y, Bae GU, Kim SN, Kim YK. Protein arginine methyltransferases: promising targets for cancer therapy. Exp Mol Med. 2021;53(5):788–808. doi:10.1038/s12276-021-00613-y
137. Eram MS, Shen Y, Szewczyk M, et al. A potent, selective, and cell-active inhibitor of human type I protein arginine methyltransferases. ACS Chem Biol. 2016;11(3):772–781. doi:10.1021/acschembio.5b00839
138. Shen Y, Szewczyk MM, Eram MS, et al. Discovery of a potent, selective, and cell-active dual inhibitor of protein arginine methyltransferase 4 and protein arginine methyltransferase 6. J Med Chem. 2016;59(19):9124–9139. doi:10.1021/acs.jmedchem.6b01033
139. Mitchell LH, Drew AE, Ribich SA, et al. Aryl pyrazoles as potent inhibitors of arginine methyltransferases: identification of the first PRMT6 tool compound. ACS Med Chem Lett. 2015;6(6):655–659. doi:10.1021/acsmedchemlett.5b00071
140. Kaushik S, Liu F, Veazey KJ, et al. Genetic deletion or small-molecule inhibition of the arginine methyltransferase PRMT5 exhibit anti-tumoral activity in mouse models of MLL-rearranged AML. Leukemia. 2018;32(2):499–509. doi:10.1038/leu.2017.206
141. Vinet M, Suresh S, Maire V, et al. Protein arginine methyltransferase 5: a novel therapeutic target for triple-negative breast cancers. Cancer Med. 2019;8(5):2414–2428. doi:10.1002/cam4.2114
142. Huang T, Yang Y, Song X, et al. PRMT6 methylation of RCC1 regulates mitosis, tumorigenicity, and radiation response of glioblastoma stem cells. Mol Cell. 2021;81(6):1276–1291e9. doi:10.1016/j.molcel.2021.01.015
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.