Back to Journals » International Journal of General Medicine » Volume 15
The Glu69Asp Polymorphism of EME1 Gene is Associated with an Increased Risk of Hepatocellular Carcinoma in Guangxi Population, China
Authors Wang Y, Huang X, Su Z, He J, Zhao N, Nie L, Tang Y, Zhao H, Nong Q
Received 25 July 2022
Accepted for publication 3 October 2022
Published 18 October 2022 Volume 2022:15 Pages 7855—7866
DOI https://doi.org/10.2147/IJGM.S383261
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Youxin Wang,1,* Xinglei Huang,1,* Zhaohui Su,1,* Junquan He,1 Na Zhao,1 Liyun Nie,1 Yanmei Tang,1 Huiliu Zhao,2 Qingqing Nong1,3
1Department of Environmental Health, School of Public Health, Guangxi Medical University, Nanning, 530021, People’s Republic of China; 2Department of Clinical Laboratory, The Affiliated Tumor Hospital of Guangxi Medical University, Nanning, 530021, People’s Republic of China; 3Guangxi Colleges and Universities Key Laboratory of Prevention and Control of Highly Prevalent Diseases, Guangxi Medical University, Nanning, 530021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qingqing Nong, Department of Environmental Health, School of Public Health, Guangxi Medical University, Nanning, 530021, People’s Republic of China, Tel +86 771-5358146, Fax +86 771-5350823, Email [email protected]
Background: The dysfunction of Essential meiotic endonuclease 1 homolog 1 (EME1) can lead to genomic instability and tumorigenesis. Single nucleotide polymorphisms (SNPs) in the EME1 gene have been reported to be associated with the risk of several cancers, but its association with hepatocellular carcinoma (HCC) has not been investigated. This study aimed to determine the association between EME1 SNPs and the risk of HCC.
Methods: This study included 645 HCC patients and 649 healthy controls from a Guangxi population of Southern China, and genotyped three functional SNPs (Glu69Asp: rs3760413A>C, Ile350Thr: rs12450550T>C, and rs11868055A>G) of the EME1 gene utilizing the Agena MassARRAY platform.
Results: The rs3760413C variant genotypes (AC+CC: Glu/Asp+Asp/Asp) conferred a 1.419-fold risk of HCC compared to the AA (Glu/Glu) genotype (adjusted OR = 1.419, 95% CI = 1.017– 1.980), and the allele C increased the risk of HCC in a dose-dependent manner (Ptrend = 0.017). Moreover, the effects of the rs3760413C variant genotypes were more pronounced in individuals who drank pond/ditch water (adjusted OR = 3.956, 95% CI = 1.413– 11.076) than in those who never drank (P = 0.033). We further observed that a potential carcinogen microcystin-LR induced more DNA oxidative damages in peripheral blood mononuclear cells from the carriers of rs3760413C variant genotypes than those from the subjects with AA genotype (P = 0.006). A nomogram was also constructed combining the rs3760413A>C polymorphism and environmental risk factors for predicting HCC risk with a good discriminatory ability (concordance index = 0.892, 95% CI: 0.874– 0.911) and good calibration (mean absolute error = 0.005).
Conclusion: Our data suggest that the Glu69Asp missense polymorphism (rs3760413) of EME1 gene is associated with the risk of HCC, which may be a susceptible biomarker of HCC in the Guangxi population.
Keywords: EME1, hepatocellular carcinoma, missense polymorphism, pond/ditch water, nomogram
Introduction
Liver cancer is the sixth most commonly diagnosed cancer and the third leading cause of cancer-related deaths worldwide.1 China is a major contributor to the global burden of liver cancer, accounting for approximately half of cases and deaths worldwide.2 The most frequent type of liver cancer is hepatocellular carcinoma (HCC), which accounts for 75–85% of all cases. The etiology of HCC is multifactorial and complex, and epidemiological studies of HCC have established many etiologic factors including alcohol, hepatitis B virus (HBV), hepatitis C virus (HCV), aflatoxin B1, microcystin-LR (MC-LR), and various carcinogens.3–5 HBV infection, exposure to aflatoxin B1 or microcystins (MCs)-contaminated drinking water has been reported to contribute to the unusually high incidence of HCC in the Guangxi Province of China.6,7 These factors can potentially modify the DNA structure and cause genomic instability, thus leading to a high risk of HCC.8,9 Efficient and accurate repairs of DNA damage could maintain the stability of the genome. However, deficiencies in the repair of DNA double-strand breaks (DSBs) might sensitize carcinogens, which can easily induce chromosomal aberrations, lead to genomic instability, and ultimately cause cancer development.10
In humans, there are two different pathways in the repair of DSBs, including the homologous recombination (HR) and non-homologous end-joining (NHEJ) pathways.11 HR is the main pathway involved in the repair of DSBs caused by the replication machinery in eukaryotes, which synthesizes across a single-strand break or an unrepaired lesion, leading to the collapse of the replication fork through several DNA repair molecules, such as essential meiotic endonuclease 1 homolog 1 (EME1). EME1 is an important member of DNA repair genes, and its encoded protein is involved in the HR repair process of DSBs. EME1 protein binds to methyl methanesulfonate-sensitive UV-sensitive 81 (MUS81) protein to form a heterodimeric complex (MUS81-EME1), which is a structure-selective endonuclease that plays a critical role in the resolution of recombination intermediates during DNA repair after inter-strand cross-links, replication fork collapse, or DNA DSBs.12–14
The human EME1 gene is located on chromosome 17q21.33 and contains 9 exons with a total length of 20 kb. This gene encodes a protein consisting of 583 amino acids with a full length of 65 kDa.15 The EME1 protein consists of a central nuclease domain, two repeated C-terminal helix–hairpin–helix (HhH) motifs, a connecting helix, and a 36-residue flexible domain linker composition, which is essential for DNA recognition.16 Currently, it is believed that the differences in the DNA repair ability of different individuals are determined by genetics, probably due to the combined effects of single nuclear polymorphisms (SNPs) of some core genes in the DNA repair pathway and environmental factors. Recent studies found that the SNPs in EME1 were associated with elevated risk of glioblastoma17 and pediatric brain tumors.18 Most of the analyses focused on a common missense polymorphism resulting in the amino acid change (isoleucine to threonine, Ile350Thr). Zhao et al reported that the exon variant Ile350Thr of EME1 was significantly associated with an increased risk of early onset of breast cancer in southern Chinese women.19 However, no study has yet tested the associations between genetic variants in EME1 gene and the risk of HCC.
In this study, we performed a case-control study that included a Chinese Han or Zhuang population of 645 HCC cases and 649 cancer-free controls to investigate the association between three SNPs (Glu69Asp: rs3760413A>C, Ile350Thr: rs12450550T>C, and rs11868055A>G) of the EME1 gene and the risk of HCC in the Guangxi Province of China. Furthermore, we also developed a risk prediction nomogram of HCC based on EME1 SNPs and environmental risk factors.
Materials and Methods
Study Subjects
A total of 645 patients with histopathologically proven HCC confirmed by at least two pathologists (Mean age: 50.1 ± 10.5 years old; 547 males and 98 females) were recruited between 2018 and 2021 in the Affiliated Tumor Hospital of Guangxi Medical University, and 649 sex and age frequency-matched cancer-free individuals (mean age: 49.2 ± 10.6 years old; 541 males and 108 females) were enrolled as a control group in the same period. The patients were all newly diagnosed with HCC, but had not been treated with radiation, chemotherapy, or surgical therapy before enrollment in the study. All the participants were ethnically Han or Zhuang Chinese who have lived in Guangxi for more than 10 years and were excluded from this study if they were affected by other types of cancer. All participants signed a written informed consent, then donated 5 mL of blood, and were interviewed using a structured questionnaire to collect their personal information, such as alcohol drinking, tobacco smoking, family history of cancer, drinking pond/ditch water, HBV and HCV infection, amongst others. After the survey, to ensure the authenticity and accuracy of the information collected by the questionnaires, this study arranged for reviewers to randomly select 5% of the participants from the survey questionnaires for a questionnaire return visit. The results of the return visit were basically the same as those of the previous questionnaire. Participants who had cumulatively smoked at least 100 cigarettes in their lifetime were considered as smokers, otherwise as non-smokers.21 Similarly, participants who had consumed alcohol at least once a week for more than one year were defined as drinkers of alcohol, otherwise as non-drinkers of alcohol.20,21 A pond/ditch was defined as a water storage place, approximately 5–6 m in width, 100 m in length, and 1–2 m in depth, usually located close to residential areas.6 Participants who had used water from ponds/ditches as the source of drinking water for 20 years or more were defined as drinkers of pond/ditch water.6 This study was approved by the Medical Ethics Committee of Guangxi Medical University (GXMU-20160303-9), and the study was conducted in accordance with the Declaration of Helsinki.
SNP Selection
By comprehensively considering the functional regions where the SNPs are located, potential biological functions, the Minor Allele Frequency (MAF), and reviewing existing studies in the literature, three putative functional SNPs related to the DNA repair gene EME1 were finally selected. After searching the NCBI dbSNP database (http://www.ncbi.nlm.nih. gov/), they were found to all have a MAF > 5% in the Chinese Han population. These three SNPs are rs11868055A>G, located in the 2000 bp upstream promoter of the EME1 gene, and Glu69Asp and Ile350Thr, located in the coding region of the EME1 gene.
Genotype Analysis
The EME1 polymorphisms were genotyped by using a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometer, and the design of primers for PCR and extension was performed using the software MassARRAY Assay Design 3.1 (Agena Bioscience, San Diego, CA). For rs3760413A>C, genomic DNA was amplified using the primers 5’-ACGTA CGAAC ACACG CACAC TTACA CACAC-3’ (forward) and 5’-ACGTA CGAAC AGATA CACAA GCCTC CACTC-3’(reverse), and the single-base extension reaction was promoted using the UEP primer 5’-GGGTT TACAC ACACA GTTTC AGCTA T-3’. For rs12450550T>C, the primers were 5’-ACGTA CGAAC CACAA TCACC AGACA CAGAG-3’ (forward) and 5’-ACGTA CGAAC AAGGG AAGGA AACGC TTCAG-3’(reverse), and the UEP primer in the single-base extension reaction was 5’- CCCTC ACCAC TCTTA CCACA C-3’. For rs11868055A>G, the primers were 5’-ACGTA CGAAC CTTAC CTCTT CACCC TACTC-3’ (forward) and 5’-ACGTA CGAAC GGTTT CACCC TAAGC AACAC-3’(reverse), and the UEP primer in the single-base extension reaction was 5’-TCTCC TCCTC AAAGT AAAAC GT-3’. The specific method has been described previously.6
Bioinformatic Analysis
To support the biological plausibility of the variant, we performed bioinformatics analysis with the UniProt database (https://www.uniprot.org/uniprot/Q96AY2) to explore the potential function of the SNP in the EME1 gene. Also, expression quantitative trait loci (eQTL) were checked with Genotype-Tissue Expression (GTEx) project (https://gtexportal.org/home/), and certain tissues were selected to present normalized effect size and P value.
Detection of Oxidative DNA Damage
To further explore the contents of DNA damage caused by MC-LR in individuals with different EME1 rs3760413 genotypes, we determined the basal and MC-LR-induced DNA oxidative damage by 8-hydroxy-2’-deoxyguanosine (8-OHdG) analysis. An additional 150 control participants were recruited to donate 6 mL of peripheral blood for isolating peripheral blood mononuclear cells (PBMCs), which were subsequently used to evaluate MC-LR exposure sensitivities. The experimental process of PBMCs isolation has been described in a previous study.6 The PBMCs were treated with 3 μg/mL MC-LR (treated group) or medium alone (untreated group) for 24 h, and then the content of 8-OHdG was assayed by the kits obtained from Nanjing Jiancheng Biology Engineering Institute (Jiangsu, China). The exact 8-OHdG value of each participant was expressed as the 8-OHdG value of MC-LR-treated group minus the 8-OHdG value of the untreated group.
Statistical Analysis
The Hardy-Weinberg equilibrium (HWE) was tested by a goodness-of-fit chi-square test to compare the expected genotype frequencies with observed genotype frequencies in controls. Student’s t-test and chi-square (χ2) tests were performed on selected variables in the case and control groups to determine their significance. Potential associations between genotype and HCC risk were examined by unconditional logistic regression under respective codominant, dominant, and recessive genetic models. Crude and adjusted odds ratios (ORs) and their 95% confidence intervals (CI) were analyzed by unconditional logistic regression unadjusted or adjusted for age, sex, ethnicity, family history of cancer, tobacco smoking, alcohol drinking, HBV infection, and drinking pond/ditch water. The logistic model was also used for the trend test of variant genotypes of the EME1. Student’s t-test and one-way analysis of variance (ANOVA) test were used to examine the difference in the levels of 8-OHdG between the different subgroups. Data analysis was performed using SPSS version 23.0 (IBM, Chicago, USA) software and GraphPad Prism 7 (GraphPad Software Inc., California, USA). Furthermore, the sample size and statistical power were calculated using the PASS software (https://www.ncss.com). The heterogeneity of different subgroups was calculated by Stata 14.0 software (StataCorp, Texas, USA). A two-sided P value < 0.05 was considered statistically significant.
Selected variables were incorporated in the nomogram to predict the probability of HCC risk using statistical software (rms in R, version 4.1.3; http://www.r-project.org). For allocating points in the nomogram, regression coefficients were applied to each individual observation to define the linear predictor. The model performance was evaluated based on the predictive accuracy for individual outcomes (discriminating ability) and the accuracy of point estimates (calibration). The model was validated using bootstrapped resampling.
Results
Basic Characteristics of Study Subjects
The basic characteristics of the 645 HCC cases and 649 healthy controls are shown in Table 1. There were no significant differences in the distribution of age, sex, ethnicity, Body Mass Index (BMI), tobacco smoking, alcohol drinking, HCV infection, and history of diabetes between cases and controls (P > 0.05 for all). However, the proportions for family history of cancer, HBsAg, HBeAg, HBV-DNA and drinking pond/ditch water were significantly different between the cases and controls (P < 0.001 for all).
 |
Table 1 Demographic and Selected Variables in HCC Cases and Controls |
Distributions of the Genotypes of EME1 SNPs and Their Risks with HCC
To determine whether the rs3760413, rs12450550, and rs11868055 polymorphisms of the EME1 gene are associated with HCC, we examined the genotypic distribution of these three polymorphisms of EME1 in both cases and controls. As shown in Table 2, the genotype frequencies of the three SNPs among controls are all in agreement with the Hardy-Weinberg equilibrium (P > 0.05 for all). The chi-square test showed that the genotype distribution of rs3760413 was significantly different between the cases and controls (P = 0.019). However, for the other two SNPs, no significant differences in genotype distribution were found (P > 0.05 for all). We determined the association of different genotype models of the three EME1 SNPs with HCC risk. Under the codominant model, the logistic regression analysis showed that the rs3760413CC (Asp/Asp) genotype conferred a 2.206-fold risk of HCC compared to the common AA (Glu/Glu) genotype (adjusted OR = 2.206, 95% CI = 1.046–4.654, P = 0.038), but not the AC (Glu/Asp) genotype (adjusted OR = 1.321, 95% CI = 0.930–1.876, P = 0.120). Meanwhile, the trend analysis indicated that the detrimental effect of EME1 rs3760413 to increase HCC risk was exhibited in a C allele dose-dependent manner (Ptrend = 0.017). Moreover, under the dominant model, the rs3760413C variant genotypes (AC+CC) increased the HCC risk by 41.9% when compared to the AA genotype (adjusted OR = 1.419, 95% CI = 1.017–1.980, P = 0.039). However, under the recessive model, no significant association with HCC risk was found (CC vs AA/AC: adjusted OR = 2.028, 95% CI = 0.969–4.242, P = 0.061). In addition, no statistical correlations were found in the other two SNPs under any genetic models.
 |
Table 2 Frequency Distribution of Genotypes in EME1 SNPs and Results of Logistic Regression Analysis for Their Associations with HCC Risk |
Stratification Analysis of the Association Between EME1 rs3760413 Variant Genotypes and HCC Risk
We performed stratified analysis to evaluate the effects of other factors on associations between EME1 genotypes and HCC risk. According to the smallest Akaike’s information criterion (AIC), the effect of rs3760413A>C best fitted the dominant model, and only the result of rs3760413A>C was presented because the other SNPs had no significant findings. As shown in Table 3, the increased HCC risk caused by the rs3760413C variant genotypes (AC+CC) was more pronounced among individuals who were drinking pond/ditch water (adjusted OR = 3.956, 95% CI = 1.413–11.076) than those who were not (Pheterogeneity = 0.033). In contrast, there were no significant differences in the stratified ORs according to sex, age, ethnicity, tobacco smoking, alcohol drinking, HBsAg, and family history of cancer (P > 0.05 for all).
 |
Table 3 Stratified Analysis of the EME1 rs3760413 A>C Genotypes by Selected Variables in HCC Cases and Controls |
Bioinformatic Analysis of the Potential Function of EME1 rs3760413A>C Polymorphism
By the use of UniProt database (https://www.uniprot.org/uniprot/Q96AY2), we found that the rs3760413A>C is a functional missense polymorphism, which causes an amino acid change from glutamic acid (Glu) to aspartic acid (Asp) at codon 69 (Glu69Asp). The SNP of rs3760413 was further selected to explore the expression quantitative trait loci (eQTL) in GETx (https://gtexportal.org/home/). As shown in Table 4, the SNP of rs3760413 mainly cis-regulated the expression of EME1 and C allele reduced the expression of EME1 gene.
 |
Table 4 Cis-Acting Genes with Transcript Level of SNPs of EME1 Gene in Selected Human Tissues |
Effects of the EME1 rs3760413A>C Polymorphism on the MC-LR-Induced Oxidative DNA Damage in PBMCs
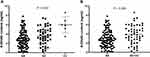
We recruited 150 additional controls and then investigated whether this rs3760413 polymorphism could affect the responses of PBMCs against MC-LR-induced DNA oxidative damage. As shown in Figure 1, the average level of 8-OHdG in the 6 cases of rs3760413CC carriers was significantly higher than that of 97 cases with AA genotype or 47 cases with AC genotype (ANOVA test: P = 0.001). Furthermore, the average level of 8-OHdG of the rs3760413C variant genotypes (AC+CC) carriers was much higher than that of AA genotype (Student’s t-test: P = 0.006).
Predictive Nomogram in HCC Risk Based on rs3760413A>C Polymorphism and Environmental Risk Factors
A nomogram calculator is presented in Figure 2. Through univariate analysis and comprehensive consideration of the risk factors of HCC, all the selected parameters (age, sex, ethnicity, rs3760413A>C polymorphism, tobacco smoking, alcohol drinking, HBsAg, family history of cancer, drinking pond/ditch water) were used to develop an individualized risk score to identify HCC. As shown in Figure 2A, higher total points based on the sum of the assigned number of points for each factor in the nomograms were associated with a higher HCC risk. The nomogram demonstrated good accuracy in estimating the risk of HCC, with a concordance index of 0.892 (95% CI: 0.874–0.911), as is displayed in Figure 2B. The calibration curve for the probability of HCC showed good agreement between the prediction of the nomogram and actual observation (mean absolute error = 0.005), and was confirmed through bootstrapping validation (Figure 2C).
Discussion
In the current hospital-based case-control study of 645 patients and 649 controls, we analyzed the associations between three functional SNPs (rs3760413A>C, rs12450550T>C, and rs11868055A>G) of the EME1 gene and HCC risk. We found that the rs3760413 polymorphism conferred a significantly increased risk of HCC in a C allele dose-dependent manner. The increased risk of HCC was more evident in participants who were drinking pond/ditch water. In contrast, we did not observe significant associations between any other two SNPs in EME1 and HCC risk. To our knowledge, this is the first study to assess the association between EME1 SNPs and HCC risk, especially in individuals who used water from ponds or ditches as the source of drinking water.
EME1 is an essential participator of the HR pathway by being part of the MUS81-EME1, which is essential for the recognition of DNA and DNA DSBs. Deficiency of the EME1 can lead to genomic instability and promote tumorigenesis in the early stage of tumor initiation.22 Several studies have reported the associations between the EME1 exon variant rs12450550T>C (Ile350Thr) and cancer risks. Adel Fahmideh et al found that EME1 rs12450550 polymorphism confers an increased risk of non-astrocytoma.18 In addition, Chang et al revealed that EME1 rs12450550 was potentially associated with glioblastoma multiforme susceptibility in the Caucasian population.17 However, EME1 rs12450550T>C is not always associated with cancer risk. For example, a study found that the rs12450550 variant genotype was not associated with the risk of glioblastoma multiforme in an Arab Jordanian population.23 Similarly, our present results also showed that this rs12450550 polymorphism was not associated with HCC risk in the Guangxi population of southern China. The reason for such discrepancies in results may be that our study is only related to Han and Zhuang Chinese.
Interestingly, we revealed that a novel SNP, rs3760413A>C, is associated with HCC risk in a Guangxi population. This rs3760413 locus is in the N-terminal region of EME1. We used UniProt database (https://www.uniprot.org/uniprot/Q96AY2) to explore the potential function of the rs3760413, and found that this functional missense polymorphism causes an amino acid change from glutamic acid to aspartic acid at codon 69. Bioinformatics analysis with the Genotype-Tissue Expression (GTEx) project (https://gtexportal.org/home/) showed that the SNP of rs3760413 mainly cis-regulated the expression of EME1 and C allele reduced the expression of EME1, which may result in reduced DNA repair capacity of EME1. This is consistent with our observation of rs3760413C variant genotypes with increased the risk of HCC. After being exposed to environmental carcinogens that damage DNA, the rs3760413C variant genotypes (AC+CC) carriers may exert weaker DNA repair capacity than AA genotype carriers, making them more susceptible to HCC.
In addition, the SNP rs3760413A>C can interact with drinking pond/ditch water to increase HCC risk as that the adverse effect of the rs3760413 variant genotypes was more pronounced in individuals with had ever drunk pond-ditch water. Epidemiological studies have reported that the high incidence of HCC in Guangxi province was partially attributed to the high consumption of pond/ditch water, and much higher levels of MC-LR were observed in pond/ditch water than in tap water.6,24–26 MC-LR is the most commonly encountered microcystin with strong hepatotoxicity and tumor promotion activity.27 Several studies have demonstrated that MC-LR can cause DNA oxidative damage in human cells and promote cancer occurrence.28–31 In the present study, we treated PBMCs with MC-LR and found that the cells from carriers of the rs3760413C variant genotypes (AC+CC) were found to induce more DNA oxidative damages than those from the AA genotype carriers, which further supports the result of this study that the EME1 rs3760413C variants may increase susceptibility to DNA oxidative damage from MC-LR. Thus, our novel finding of gene-environment interaction between the EME1 variant genotypes and drinking pond/ditch water on increasing cancer risk is biologically acceptable We also constructed a genetic-environment interaction model to predict HCC risk. A predictive nomogram on the SNP rs3760413, drinking pond/ditch water, and other risk factors showed a better predictive capability (concordance index = 0.892, P < 0.001).
In the current study, our selection of subjects met the requirement of random sampling in that the genotype frequencies among controls fitted the Hardy-Weinberg disequilibrium. In addition, we achieved over 95% study power (two-sided test, α = 0.05) to detect an OR of 1.419 for the rs3760413C variant genotypes compared with the rs3760413AA genotype, suggesting that this finding is noteworthy. However, some limitations still need to be considered. First, the sample size of our study is relatively small. Second, some selection biases were unavoidable because this was a hospital-based case-control study, and our study subjects came only from the Chinese Han or Zhuang population. Moreover, a considerable proportion of patients lacked of clinical stages and cancer differentiation status, thus limiting our analysis on the associations between EME1 variants and clinical features.
Conclusions
In conclusion, our study found that the Glu69Asp missense polymorphism (rs3760413) of EME1 gene was significantly associated with an increased risk of HCC in Guangxi population, China. Particularly, the association was more pronounced in individuals who used water from ponds/ditches as the source of drinking water. We also constructed a nomogram model based on the Glu69Asp polymorphism and environmental risk factors that can better predict HCC. These findings all suggest that the Glu69Asp polymorphism of EME1 gene may be a susceptible biomarker for HCC in the Guangxi population.
Institutional Review Board Statement
This study was approved by the Medical Ethics Committee of Guangxi Medical University (GXMU-20160303-9). Written informed consent was obtained from all included subjects.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81960582 and No. 81660529).
Disclosure
The authors declare no conflicts of interest in relation to this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Im PK, Millwood IY, Kartsonaki C, et al. Alcohol drinking and risks of liver cancer and non-neoplastic chronic liver diseases in China: a 10-year prospective study of 0.5 million adults. BMC Med. 2021;19(1):216. doi:10.1186/s12916-021-02079-1
3. Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi:10.1001/jamaoncol.2017.3055
4. Liu Z, Jiang Y, Yuan H, et al. The trends in incidence of primary liver cancer caused by specific etiologies: results from the Global Burden of Disease Study 2016 and implications for liver cancer prevention. J Hepatol. 2019;70(4):674–683.
5. Chen T, Liu J, Li Y, Wei S. Burden of disease associated with dietary exposure to aflatoxins in China in 2020. Nutrients. 2022;14(5):1027. doi:10.3390/nu14051027
6. Wang Y, Huang Q, Huang X, et al. Genetic variant of PP2A subunit gene confers an increased risk of primary liver cancer in Chinese. Pharmgenomics Pers Med. 2021;14:1565–1574. doi:10.2147/PGPM.S335555
7. Xiang X, Qin HG, You XM, et al. Expression of P62 in hepatocellular carcinoma involving hepatitis B virus infection and aflatoxin B1 exposure. Cancer Med. 2017;6(10):2357–2369. doi:10.1002/cam4.1176
8. Long XD, Zhao D, Wang C, et al. Genetic polymorphisms in DNA repair genes XRCC4 and XRCC5 and aflatoxin B1-related hepatocellular carcinoma. Epidemiology. 2013;24(5):671–681. doi:10.1097/EDE.0b013e31829d2744
9. Díez-Quijada L, Hercog K, Štampar M, et al. Genotoxic effects of cylindrospermopsin, microcystin-LR and their binary mixture in human Hepatocellular Carcinoma (HepG2) cell line. Toxins. 2020;12:12.
10. Ui A, Chiba N, Yasui A. Relationship among DNA double-strand break (DSB), DSB repair, and transcription prevents genome instability and cancer. Cancer Sci. 2020;111(5):1443–1451. doi:10.1111/cas.14404
11. Ensminger M, Löbrich M. One end to rule them all: non-homologous end-joining and homologous recombination at DNA double-strand breaks. Br J Radiol. 2020;93(1115):20191054. doi:10.1259/bjr.20191054
12. Hanada K, Budzowska M, Modesti M, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25(20):4921–4932. doi:10.1038/sj.emboj.7601344
13. Hope JC, Cruzata LD, Duvshani A, Mitsumoto J, Maftahi M, Freyer GA. Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe. Mol Cell Biol. 2007;27(10):3828–3838. doi:10.1128/MCB.01596-06
14. Gwon GH, Jo A, Baek K, et al. Crystal structures of the structure-selective nuclease Mus81-Eme1 bound to flap DNA substrates. EMBO J. 2014;33(9):1061–1072. doi:10.1002/embj.201487820
15. Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J Biol Chem. 2003;278(27):25172–25178. doi:10.1074/jbc.M302882200
16. Chang JH, Kim JJ, Choi JM, Lee JH, Cho Y. Crystal structure of the Mus81-Eme1 complex. Genes Dev. 2008;22(8):1093–1106.
17. Chang JS, Yeh RF, Wiencke JK, et al. Pathway analysis of single-nucleotide polymorphisms potentially associated with glioblastoma multiforme susceptibility using random forests. Cancer Epidemiol Biomarkers Prev. 2008;17(6):1368–1373. doi:10.1158/1055-9965.EPI-07-2830
18. Adel Fahmideh M, Lavebratt C, Schüz J, et al. Common genetic variations in cell cycle and DNA repair pathways associated with pediatric brain tumor susceptibility. Oncotarget. 2016;7(39):63640–63650. doi:10.18632/oncotarget.11575
19. Zhao J, Liu L, Zhang A, et al. Effect of EME1 exon variant Ile350Thr on risk and early onset of breast cancer in southern Chinese women. J Biomed Res. 2013;27(3):193–201. doi:10.7555/JBR.27.20130013
20. Petrick JL, Campbell PT, Koshiol J, et al. Tobacco, alcohol use and risk of hepatocellular carcinoma and intrahepatic cholangiocarcinoma: the liver cancer pooling project. Br J Cancer. 2018;118(7):1005–1012. doi:10.1038/s41416-018-0007-z
21. Millwood IY, Li L, Smith M, et al. Alcohol consumption in 0.5 million people from 10 diverse regions of China: prevalence, patterns and socio-demographic and health-related correlates. Int J Epidemiol. 2013;42(3):816–827. doi:10.1093/ije/dyt078
22. Abraham J, Lemmers B, Hande MP, et al. Eme1 is involved in DNA damage processing and maintenance of genomic stability in mammalian cells. EMBO J. 2003;22(22):6137–6147. doi:10.1093/emboj/cdg580
23. Al-Khatib SM, Abdo N, Al-Eitan LN, et al. The impact of the genetic polymorphism in DNA repair pathways on increased risk of glioblastoma multiforme in the Arab Jordanian population: a case-control study. Appl Clin Genet. 2020;13:115–126. doi:10.2147/TACG.S248994
24. Duy TN, Lam PK, Shaw GR, Connell DW. Toxicology and risk assessment of freshwater cyanobacterial (blue-green algal) toxins in water. Rev Environ Contam Toxicol. 2000;163:113–185. doi:10.1007/978-1-4757-6429-1_3
25. Ueno Y, Nagata S, Tsutsumi T, et al. Detection of microcystins, a blue-green algal hepatotoxin, in drinking water sampled in Haimen and Fusui, endemic areas of primary liver cancer in China, by highly sensitive immunoassay. Carcinogenesis. 1996;17(6):1317–1321. doi:10.1093/carcin/17.6.1317
26. Xiao CC, Chen MJ, Mei FB, et al. Influencing factors and health risk assessment of microcystins in the Yongjiang river (China) by Monte Carlo simulation. Peer J. 2018;6:e5955. doi:10.7717/peerj.5955
27. Zegura B. An overview of the mechanisms of microcystin-LR genotoxicity and potential carcinogenicity. Mini Rev Med Chem. 2016;16(13):1042–1062. doi:10.2174/1389557516666160308141549
28. Chen L, Yang S, Wen C, et al. Regulation of microcystin-LR-Induced DNA damage by miR-451a in HL7702 cells. Toxins. 2019;11(3):164.
29. Liu W, Wang L, Zheng C, et al. Microcystin-LR increases genotoxicity induced by aflatoxin B1 through oxidative stress and DNA base excision repair genes in human hepatic cell lines. Environ Pollut. 2018;233:455–463. doi:10.1016/j.envpol.2017.10.067
30. Jia X, Guan B, Liao J, et al. Down-regulation of GCLC is involved in microcystin-LR-induced malignant transformation of human liver cells. Toxicology. 2019;421:49–58. doi:10.1016/j.tox.2019.03.010
31. Wen C, Zheng S, Yang Y, et al. Effects of microcystins-LR on genotoxic responses in human intestinal epithelial cells (NCM460). J Toxicol Environ Health. 2019;82(21):1113–1119. doi:10.1080/15287394.2019.1698498
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.