Back to Journals » Infection and Drug Resistance » Volume 16
The First Report of Escherichia coli and Klebsiella pneumoniae Strains That Produce Both NDM-5 and OXA-181 in Jiangsu Province, China
Received 28 March 2023
Accepted for publication 18 May 2023
Published 24 May 2023 Volume 2023:16 Pages 3245—3255
DOI https://doi.org/10.2147/IDR.S412678
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Guixiang Tao,1,* Hua Tan,2,* Qian Chen1
1Institute of Pediatrics, Children’s Hospital of Nanjing Medical University, Nanjing, People’s Republic of China; 2Department of Clinical Laboratory, Children’s Hospital of Nanjing Medical University, Nanjing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Qian Chen, Institute of Pediatrics, Children’s Hospital of Nanjing Medical University, Nanjing, People’s Republic of China, Tel +8618951768213, Email [email protected]
Objective: The aim of this study was to analyze the genetic characteristics of three Enterobacteriaceae strains (one strain of Escherichia coli and two strains of Klebsiella pneumoniae) that produce both the NDM-5 and OXA-181 carbapenemases in pediatric patients.
Methods: Carbapenem-resistant Enterobacteriaceae (CRE) strains were collected from the Children’s Hospital Affiliated to Nanjing Medical University in 2022. Resistance genes were detected by PCR. CRE strains that produced both the blaNDM-5 and blaOXA-181 genes were further characterized by antimicrobial susceptibility testing, multilocus sequence typing (MLST), plasmid conjugation assay, S1 nuclease-PFGE, Southern blotting and whole-genome sequencing.
Results: Three Enterobacteriaceae strains carrying both the blaNDM-5 and blaOXA-181 resistance genes were screened. MLST results showed that the strain of Escherichia coli carrying both blaNDM-5 and blaOXA-181 was ST410; the two strains of Klebsiella pneumoniae with both blaNDM-5 and blaOXA-181 were ST2601 and ST759. Conjugation assays showed that the plasmids harboring the blaNDM-5 and blaOXA-181 genes were self-transmissible. S1-PFGE and Southern blotting showed that the blaNDM-5 and blaOXA-181 genes were located on the plasmid with the size of about 60kb~. The genotyping results showed that the plasmid types were ColKP3 and IncX3.
Conclusion: This is the first report of Enterobacteriaceae strains that produce both NDM-5 and OXA-181 isolated from pediatric patients in China. Active infection control measures are urgently needed to prevent the spread of bacteria in children.
Keywords: Escherichia coli, Klebsiella pneumoniae, NDM-5, OXA-181, plasmid
Introduction
In recent years, the continuous emergence of carbapenem-resistant Enterobacteriaceae (CRE) strains threatens public health worldwide. The main mechanism of carbapenem resistance in Enterobacteriaceae is the production of KPC (Ambler class A), NDM, VIM, IMP (class B) and OXA-48-like (class D) β-lactamases. New Delhi gold β-lactamase (NDM-1) was first discovered in 2008,1 and since then, Enterobacteriaceae carrying the blaNDM gene have been found around the world. To date, 31 gene subtypes have been found globally.2 In 2011, Hornsey et al first identified blaNDM-5 in a strain of Escherichia coli ST648 with a multidrug resistance phenotype in the UK.3 Since then, strains carrying blaNDM-5 have been found in many countries around the world, including Egypt,4 South Korea,5 Italy6 and China.7 Compared with NDM-1, the NDM-5 enzyme has two amino acid substitutions (Val88Leu and Met154Leu), and it has been shown that the NDM-5 enzyme causes a higher level of resistance to carbapenems and broad-spectrum cephalosporins.3
Since 2004, when Klebsiella pneumoniae carrying blaOXA-48was isolated from a patient living in Turkey,8 Enterobacteriaceae strains carrying blaOXA-48-like have been found around the world. Several OXA-48-like carbapenemases have been reported, including OXA-48, OXA-162, OXA-181, OXA-204, OXA-232, OXA-244, and OXA-245.9 OXA-181, a variant of OXA-48 with four amino acid substitutions, was first reported in India in 2007.10 Since then, it has been found mainly in Escherichia coli and Klebsiella pneumoniae strains in several countries (UK, USA, etc.). The gene encoding OXA-181 is usually located on the IncX3-type plasmid.11 This plasmid carries several carbapenemase genes, including blaKPC12 and blaNDM.13 To date, OXA-181-producing Escherichia coli has been reported in Sichuan14 and Henan,15 and OXA-181-producing Klebsiella pneumoniae has been reported in Zhejiang.16 The first strain containing blaOXA-48-like and blaNDM- was reported in Singapore in 2013, followed by Egypt,17 the United States,18 Bangladesh,19 Italy20 and other locations. Here, we report the first Escherichia coli and Klebsiella pneumoniae strains in China that produce both the NDM-5 and OXA-181 enzymes to analyze the genetic characteristics and environment in pediatric patients.
Materials and Methods
Clinical Data
On January 6, 2022, a 24-day-old newborn with “poor response, abdominal distension” was admitted to the children’s hospital of Nanjing Medical University. After admission, abdominal distension did not improve after symptomatic treatments, including fasting, anti-infection and defecation, so an emergency exploratory laparotomy was performed that night. Necrotizing enterocolitis was confirmed during the operation, and most of the small intestine was necrotic. “Intestinal resection, jejunostomy and intestinal adhesiolysis” were performed; the operation was successful, and the treatment was continued in the neonatal Medical Center after the operation. The patient was diagnosed with neonatal necrotizing enterocolitis, peritonitis and neonatal sepsis and was given meropenem combined with vancomycin to prevent infection. Sputum cultures and drug sensitivity testing on January 8 revealed that the identified Escherichia coli (EC73) strain was multidrug resistant and only sensitive to amikacin, polymyxin and tigecycline. The infant was placed in the isolation room for MDRO isolation. On January 10, vancomycin was discontinued, and immunoglobulin was given as a supportive therapy. On March 28, the infant had fever with a peak of 40.5°C and some fatigue. On March 30, a blood-bacterial culture and drug sensitivity testing revealed that there were gram-negative bacilli present. On April 1, blood-bacterial culture and drug sensitivity assays revealed that the identified Klebsiella pneumoniae (KP92) strain was sensitive to aztreonam. Meropenem was discontinued, and the anti-bacterial treatment aztreonam was given. After 8 days, the blood cultures were negative for CRE.
On April 24, the third strain (KP100) was extracted from the abdominal effusion of an 8-month-old child three days after enterostomy. As the drug sensitivity results showed sensitivity to aztreonam, the child was given this treatment as an anti-infective. Neither of the patients had traveled in the 30 days before their admission.
Antimicrobial Susceptibility Testing and Identification of Antibiotic Resistance Genes
This study was conducted at the Children’s Hospital of Nanjing Medical University in Jiangsu Province, China. Enterobacteriaceae was identified by a VITEK-2 Compact system (bioMerieux, Marcy-L ‘Etoile, France). Antimicrobial susceptibility testing was performed by broth microdilution. Susceptibility breakpoints were set according to criteria from the Institute of Clinical and Laboratory Standards (CLSI),21 except for polymyxin E and tigecycline, whose susceptibility breakpoints were set based on criteria from the European Committee for Antimicrobial Susceptibility Testing (EUCAST) 10.0.22 The quality control (QC) strain Escherichia coli ATCC 25922 was used in all tests.
PCR was used to identify blaNDM-5 and blaOXA-181 using the following primers previously described in the literature: NDM-5-F, 5’-GAAGCTGAGCACCGCATTAG-3’, NDM-5-R, 5’-GGGCCGTATGAGTGATTGC-3’,23 OXA-48-like-F, 5ʹ-GCGTGGTTAAGGATGAACAC-3ʹ and OXA-48-like-R, 5ʹ- CATCAAGTTCAACCCAACCG −3ʹ.24
The amplification system consisted of 50 μL of ddH2O (20 μL), Multiplex Buffer (25 μL), Primer1 (10 μM) (2 μL), Primer2 (10 μM) (2 μL), and template (1 μL). The PCR amplification conditions were 95 °C for 3 min; 35 cycles of 95 °C for 15s, 55°C for 15s, and 72°C for 15s; and 72 °C for 5 min. The PCR products were identified using a 1.0% agarose gel for electrophoresis, and the positive products were sequenced by Sanger sequencing. The complete sequences were compared with those reported in GenBank using the BLAST algorithm (http://www.ncbi.nlm.nih.gov/blast/). Enterobacteriaceae strains carrying both blaNDM-5 and blaOXA-181 were collected for further study.
Multilocus Sequence Typing (MLST)
Referring to the Pasteur MLST website (http://bigsdb.Pasteur.fr/), seven housekeeping genes from Klebsiella pneumoniae, rpoB, infB, phoE, mdh, pgi, gapA, and tonB, and seven housekeeping genes from Escherichia coli, adk, fumC, gyrB, icd, mdh, purA, and recA, were amplified. DNA sequencing was performed on the positive products. The allelic profiles of the seven housekeeping genes for the respective strains were obtained from the MLST database and then submitted to the MLST website to determine the sequence typing of clinical isolates.
Conjugation Assay
Conjugation assays were performed to test the ability of plasmids carrying blaNDM-5 and blaOXA-181 to transfer between strains, and the rifampicin-resistant E. coli strain EC600 was used as the receptor. Overnight cultures of donor and recipient strains were mixed and dropped on sterile filter paper at a 1:3 ratio and incubated overnight at 37°C on MH agar plates. blaNDM-5 and blaOXA-181 conjugates were selected by screening on dual antibody plates containing imipenem (4mg/L) and rifampicin (600 mg/L).
S1-PFGE and Southern Blotting
Drug-resistant plasmids carrying the blaNDM-5 and blaOXA-181 genes were isolated and mapped by S1-PFGE and Southern blotting. Briefly, isolates with blaNDM-5 and blaOXA-181 were embedded in gold agarose and digested with S1 nuclease (TaKaRa Biotechnology Co., Dalian, China), and plasmid DNA (Bio-Rad, USA) was analyzed by PFGE electrophoresis. Then, DNA fragments were transferred to positively charged nylon membranes and hybridized specifically with blaNDM-5 and blaOXA-181 probes. Probes were synthesized using the DIG High Prime DNA Labeling and Detection Starter Kit I (Roche Applied Sciences, Penzberg, Germany) according to the manufacturer’s instructions.
Whole-Genome Sequencing and Analysis
To get a comprehensive picture of the strains that produced both blaNDM-5 and blaOXA-181, the three strains that were identified in this study (EC73, KP92 and KP100) were selected for whole-genome sequencing. Genome sequencing was performed by the Personal Biotechnology Company (Shanghai, China) using the Pacific Biosciences platform and the Illumina MiSeq/Hiseq/Novaseq platform.
The whole-genome shotgun (WGS) strategy and next-generation sequencing (NGS) technology were used to construct libraries of different insertions. The libraries were sequenced on the PacBio Sequel sequencing platform which is based on the Illumina NovaSeq sequencing platform and third-generation single-molecule sequencing technology. FastQC was used for quality control. Using CARD (The Comprehensive Antibiotic Resistance Database), antibiotic resistance genes in the genome sequence can be identified through analysis of antibiotic resistance profiles. PlasmidFinder 2.1 was used to analyze plasmid replicon types. Reference sequences were searched through the NCBI blast database, comparative gene cluster analysis was performed using Easyfig, and comparative genomic circle maps were constructed using CGView.
Results
Antimicrobial Susceptibility and Resistance Gene Testing
The three CRE strains were all resistant to imipenem, meropenem, cephalosporin and enzyme inhibitor compounds, and the Escherichia coli strain was only sensitive to amikacin, polymyxin and tigecycline. Both Klebsiella pneumoniae strains were sensitive to aztreonam, amikacin, ciprofloxacin, levofloxacin, gentamicin, tobramycin, polymyxin and tigecycline. The minimum inhibitory concentration (MIC) assay results for the three CRE strains are shown in Table 1.
 |
Table 1 The Minimum Inhibitory Concentration of Donor Bacteria, Recipient Bacteria and Conjugant (MIC, μg /Ml) |
All three CRE strains were found to carry the carbapenemase resistance genes blaNDM-5 and blaOXA-181.
MLST
MLST analysis showed that the EC73 strain is ST410, the KP92 strain ST2601 and the KP100 strain ST759.
Plasmid Conjugation Assay and Gene Location
The plasmids carrying the blaNDM-5 and blaOXA-181 genes could be transferred from the donor strain to the recipient strain EC600. The corresponding conjugants are named J73, J92, and J100. The results of PCR amplification showed that all the conjugants carried the blaNDM-5 and blaOXA-181 genes. The drug sensitivity results showed that the three conjugants were resistant to carbapenems and enzyme inhibitors, as shown in Table 1.
The results of S1-PFGE and Southern blotting showed that blaNDM-5 and blaOXA-181 were located on plasmids of approximately 60 kb in size in the three donor strains and in the three conjugants, as shown in Figure 1.
Plasmid Sequence Analysis Based on Whole-Genome Sequencing
The CARD database was used to analyze the whole-genome sequencing results, and the predicted resistance genes in the EC73, KP92 and KP100 strains were identical as follows: 1) the β-lactam resistance genes: blaNDM-5 and blaOXA-181; 2) the quinolone resistance gene: qnrS1; 3) the aminoglycoside resistance gene: aadA2; 4) the sulfa resistance gene: sul; 5) the methylene benacil resistance gene: dfrA12; and 6) the quaternary ammonium resistance gene: qacEΔ1. The carbapenemase resistance gene blaOXA-181 was located on the 61,973 bp plasmid 2. The length of EC73 plasmid 2 was 61,973 bp, the average GC content was 48.06%, and there were 81 open reading frames. The length of KP92 plasmid 2 was 61,973 bp, the average GC content was 48.06%, and there were 81 open reading frames. The length of KP100 plasmid 2 was 61,973 bp, the average GC content was 48.06%, and there were 83 open reading frames. According to the PlasmidFinder database from the Center for Genomic Epidemiology, the bacterial drug resistance genes blaNDM-5 and blaOXA-181 were on the IncX3 and ColKP3 plasmids in this study. The skeleton structure of these plasmids includes the plasmid conjugation T4SS secretion system, which consists of the Vir gene family (virB2, virB3, virB9, virB10, virB11), the protein-related genes parA and topB that help maintain plasmid stability, and the traG and trbM elements that play a role in conjugation and transfer.
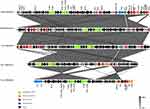
The blaNDM-5 and blaOXA-181 carrier genes are located on the Tn3 gene transposable unit. The complete sequences of the three strains (EC73, KP92 and KP100) have been deposited in GenBank, and the accession numbers are SRP399849, SRP398461 and SRP398463. Further BLAST plasmid sequence alignment analysis showed that the nucleotide similarity between EC73 plasmid 2 and NZ_CP024825.1 (Escherichia coli isolated from Korea in 2015, blaNDM-5 located on the IncX3 plasmid) was more than 99%, and KP92 plasmid 2 and NZ_CP026727.1 (Escherichia coli isolated from the United Kingdom in 2018, blaOXA-181 is located on the IncX3 plasmid) had more than 99% nucleotide similarity. The genome comparisons are shown in Figures 2 and 3.
 |
Figure 2 Comparison of NZ_CP024825.1, NZ_CP026727.1, KP100, KP92 and EC73 genomes from the outside in. |
Discussion
Carbapenems have long been considered the last line of defense for multidrug-resistant gram-negative bacterial infections. The emergence of carbapenem-resistant Klebsiella pneumoniae is a serious challenge as there are few clinical anti-infective treatments, especially in pediatric patients. The purpose of this study was to investigate the resistance-related characteristics of CRE strains isolated in the Children’s Hospital Affiliated to Nanjing Medical University that produce both the NDM-5 and OXA-181 enzymes to help prevent large-scale hospitalization of patients with CRE in the Children’s Hospital. To the best of our knowledge, this is the first time in China that Escherichia coli and Klebsiella pneumoniae strains that produce both the NDM-5 and OXA-181 enzymes have been found in children.
The two children whose samples were used in this study were both preterm infants who had very low birth weights. Since preterm birth and low birth weight are both risk factors for CRE infection in children,25 children with high risk factors should receive more attention when implementing prevention and control measures in the future. In this study, all clinical strains were sensitive to polymyxin and tigecycline in vitro. Tigecycline is not recommended for children because of the risk of tooth coloring, and polymyxin is not used for the clinical treatment of children in China.26 In addition, aminoglycoside antibiotics and quinolones are rarely used in children because of their ototoxicity and nephrotoxicity.27,28 Clinical data showed that both children had good clinical effects after treatment with aztreonam, which was consistent with the drug resistant profiles of the clinical isolates. Therefore, it is possible that the strains isolated from sputum specimens may be a part of the colonizing flora in the respiratory tract rather than true pathogens. Because this study was retrospective, we were unable to conduct an in-depth analysis of the environment surrounding the patients and the source of the strains.
According to the MLST results in this study, the EC73 strain is ST410, the KP92 strain is ST2601, and the KP100 strain is ST759. The Escherichia coli strain identified in this study is ST410, which belongs to the ST23 clonal complex and has been considered the origin of the ST23 complex. Escherichia coli strains that are ST410 are widely distributed in Europe (Poland,29 Denmark,30 Italy,31 etc.), Asia (South Korea,32 Singapore,33 India,34 China,35 etc.), Africa,36 North America (United States,37 Mexico,38 etc.) and South America (Brazil,39 Chile40 et al), etc. However, Klebsiella pneumoniae strains that are ST2601 and ST759 and that produce both NDM-5 and OXA-181, such as the strains used in this study, have never been reported globally.
The results of conjugation experiments showed that the resistance genes blaNDM-5 and blaOXA-181 in the three CRE strains could be successfully transferred into the recipient strain EC600 by plasmid, which made the conjugants resistant to carbapenems. Plasmid analysis showed that blaNDM-5 and blaOXA-181 are located on the IncX3 and ColKP3 plasmids, respectively, both of which are the same size (approximately 61 kb). The ColKP3 plasmid is not self-transmitted but can move with the help of other plasmids.9 The IncX3 plasmid plays an important role in the transmission of the blaNDM-5 gene among Enterobacteriaceae.7 IncX3-type plasmids have also been frequently reported to mediate the spread of other NDM subtypes, including blaNDM-1, blaNDM-4, blaNDM-5, blaNDM-6, blaNDM-7, blaNDM-13, blaNDM-17, blaNDM-19, blaNDM-20, and blaNDM-21.41 Therefore, we need to take effective measures to control the spread of this resistance plasmid among strains.
Whole-genome sequencing of the CRE strains revealed blaNDM-5 and blaOXA-181 to be present on the IncX3 and ColKP3 plasmids that were 61,973 bp in size. BLAST analysis of plasmid sequences showed that plasmid 2 of EC73 is highly similar to that in NZ_CP024825.1 (Escherichia coli isolated from Korea in 2015) and that plasmid 2 of KP92 is highly similar to that in NZ_CP026727.1 (Escherichia coli isolated from the UK in 2018). These findings suggest that the CRE plasmids that carry both of the carbapenemase genes may be derived from Escherichia coli. In plasmids, drug resistance genes are often associated with mobile genetic elements, such as transposons (Tn) and insertion sequences (IS). IS26 belongs to the IS6 family, which plays a pivotal role in the transmission of antibiotic resistance genes among gram-negative bacilli.42 IS26 encloses multiple resistance genes and lays the groundwork for expression of resistance genes.43 In the three CRE strains in this study, blaNDM-5 is surrounded by two IS26 on both sides to form the complex transposon in the core region of dfrA12-aadA2-qacEΔ1-sul-ISCR1-dsbD-trpF-bleMBL-blaNDM-5 (not seen in NZ_CP024825.1), and blaOXA-181 is flanked by two IS26 envelopes to form the qnrS1-blaOXA-181 resistance gene cassette (also present in NZ_CP026727.1), with the qnrS1 gene being associated with low levels of fluoroquinolone resistance. The downstream elements of the blaNDM-5 gene (bleMBL, trpF and dsbD) often appeared in various plasmids, and a large number of drug resistance genes were found to coexist upstream and downstream of NDM-5.44,45 The drug resistance genes included dfrA12, aadA2, qacEΔ1 and sul. The ISCR1 element was first reported by the Australian scholar Stokes.46 The ISCR1 element can recognize and carry downstream drug resistance genes via transposition by rolling loop replication. Because ISCR1 can easily carry drug resistance genes, it leads to the spread of drug resistance.47 In addition, the three CRE strains in this study were found to carry the disinfectant resistance gene qacEΔ1, indicating that Enterobacteriaceae strains have increased resistance to disinfectants that are commonly used in hospitals. The disinfectants commonly used in clinical practice include iodophor, Pasteurian disinfectant, 533 (chlorine-containing disinfectant), Aijiajia (chlorhexidine gluconate), chlorhexidine, and benzalkonium chloride. However, due to the widespread and unreasonable use of various disinfectants, selective pressure on bacteria causes them to become insensitive to disinfectants or makes conventionally used disinfectant concentrations ineffective. The qacEΔ1 gene encodes a transmembrane protein that is resistant to quaternary ammonium disinfectants (such as benzylammonium bromide and neogeramine). Due to the linkage of the qacEΔ1 and sul1 genes, these bacteria are resistant to sulfonamides.48 Therefore, we should switch the disinfectants that are regularly used in the clinic to minimize bacterial tolerance to disinfectants.
Although international travel has been reported to accelerate the spread of NDM and OXA-48-like enzymes, neither the children nor the mothers in this study had a history of international travel. At present, more epidemiological studies are urgently needed to better understand the emergence and transmission mechanism of the blaNDM-5 and blaOXA-181 genes in China.
Conclusion
In conclusion, this study is the first report of Escherichia coli and Klebsiella pneumoniae strains that produce carbapenemases (NDM-5 and OXA-181) in pediatric patients in Jiangsu Province, China. These strains are a major threat because they make treatment and recovery more difficult in young children. Plasmid analysis showed the blaNDM-5 and blaOXA-181 genes to be located on the IncX3 and ColKP3 plasmids with a size of 61 kb. The IncX3 plasmid, which is a carrier of drug resistance genes and is associated with a risk of spread in pediatric wards, showed a high degree of autonomous transfer between strains. Therefore, we need to actively monitor the genetics of CRE strains, especially in immunocompromised children, and take more stringent control measures to prevent the further spread of these multidrug-resistant strains in pediatric wards. In addition, clinicians need to fully understand the characteristics of disinfectant resistance genes and their current status to establish sound norms for the use of disinfectants and to reduce the spread of these resistance genes caused by irregular use or abuse of disinfectants.
Ethical Approval
The guardian of the pediatric patients signed informed consent to participate in the study before the study began and this study was conducted in accordance with the Declaration of Helsinki. The Clinical Research Ethics Committee of the Children’s Hospital of Nanjing Medical University approved the study (202205054-1), as all samples collected in this work were initially used to diagnose patient care without increasing the patient’s medical costs and suffering.
Consent for Publication
Written informed consent was provided by the patient guardian for the publication of the case details.
Funding
This research was supported by the Second Level of the Nanjing Health Youth Talents Training Project (QRX17078).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother. 2009;53(12):5046–5054. doi:10.1128/AAC.00774-09
2. Li X, Zhao D, Li W, Sun J, Zhang X. Enzyme inhibitors: the best strategy to tackle superbug NDM-1 and its variants. Int J Mol Sci. 2021;23(1):197. doi:10.3390/ijms23010197
3. Hornsey M, Phee L, Wareham DW. A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 2011;55(12):5952–5954. doi:10.1128/AAC.05108-11
4. Ramadan H, Gupta SK, Sharma P, et al. Circulation of emerging NDM-5-producing Escherichia coli among humans and dogs in Egypt. Zoonoses Public Health. 2020;67(3):324–329. doi:10.1111/zph.12676
5. Hong JS, Song W, Jeong SH. Molecular characteristics of NDM-5-producing Escherichia coli from a cat and a dog in South Korea. Microb Drug Resist. 2020;26(8):1005–1008. doi:10.1089/mdr.2019.0382
6. Alba P, Taddei R, Cordaro G, et al. Carbapenemase IncF-borne blaNDM-5 gene in the E. coli ST167 high-risk clone from canine clinical infection, Italy. Vet Microbiol. 2021;256:109045. doi:10.1016/j.vetmic.2021.109045
7. Tian D, Wang B, Zhang H, et al. Dissemination of the bla NDM-5 gene via IncX3-type plasmid among Enterobacteriaceae in children. mSphere. 2020;5(1):e00699–19. doi:10.1128/mSphere.00699-19
8. Poirel L, Heritier C, Tolun V, Nordmann P. Emergence of oxacillinase-mediated resistance to imipenem in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(1):15–22. doi:10.1128/AAC.48.1.15-22.2004
9. Pitout J, Peirano G, Kock MM, Strydom K-A, Matsumura Y. The global ascendency of OXA-48-type carbapenemases. Clin Microbiol Rev. 2019;33(1):e00102–19. doi:10.1128/CMR.00102-19
10. Castanheira M, Deshpande LM, Mathai D, Bell JM, Jones RN, Mendes RE. Early dissemination of NDM-1- and OXA-181-producing Enterobacteriaceae in Indian hospitals: report from the SENTRY Antimicrobial Surveillance Program, 2006–2007. Antimicrob Agents Chemother. 2011;55(3):1274–1278. doi:10.1128/AAC.01497-10
11. Izdebski R, Baraniak A, Zabicka D, et al. Enterobacteriaceae producing OXA-48-like carbapenemases in Poland, 2013–January 2017. J Antimicrob Chemother. 2018;73(3):620–625. doi:10.1093/jac/dkx457
12. Fuga B, Ferreira ML, Cerdeira LT, et al. Novel small IncX3 plasmid carrying the blaKPC-2 gene in high-risk Klebsiella pneumoniae ST11/CG258. Diagn Microbiol Infect Dis. 2020;96(2):114900. doi:10.1016/j.diagmicrobio.2019.114900
13. Zhu W, Wang X, Qin J, Liang W, Shen Z. Dissemination and stability of the blaNDM-5-carrying IncX3-type plasmid among multiclonal Klebsiella pneumoniae isolates. mSphere. 2020;5(6):e00917–20. doi:10.1128/mSphere.00917-20
14. Liu Y, Feng Y, Wu W, et al. First report of OXA-181-producing Escherichia coli in china and characterization of the isolate using whole-genome sequencing. Antimicrob Agents Chemother. 2015;59(8):5022–5025. doi:10.1128/AAC.00442-15
15. Qin S, Cheng J, Wang P, Feng X, Liu HM. Early emergence of OXA-181-producing Escherichia coli ST410 in China. J Glob Antimicrob Resist. 2018;15:215–218. doi:10.1016/j.jgar.2018.06.017
16. Liu C, Fang Y, Zeng Y, et al. First report of OXA-181-producing Klebsiella pneumoniae in China. Infect Drug Resist. 2020;13:995–998. doi:10.2147/IDR.S237793
17. Gamal D, Fernández-Martínez M, El-Defrawy I, Ocampo-Sosa AA, Martínez-Martínez L. First identification of NDM-5 associated with OXA-181 in Escherichia coli from Egypt. Emerg Microbes Infect. 2016;5(4):e30. doi:10.1038/emi.2016.24
18. Rojas LJ, Hujer AM, Rudin SD, et al. NDM-5 and OXA-181 beta-lactamases, a significant threat continues to spread in the Americas. Antimicrob Agents Chemother. 2017;61(7):e00454–17. doi:10.1128/AAC.00454-17
19. Okanda T, Haque A, Koshikawa T, et al. Characteristics of carbapenemase-producing Klebsiella pneumoniae isolated in the intensive care unit of the largest tertiary hospital in Bangladesh. Front Microbiol. 2021;11:612020. doi:10.3389/fmicb.2020.612020
20. Marchetti VM, Bitar I, Mercato A, et al. Complete nucleotide sequence of plasmids of two Escherichia coli strains carrying blaNDM–5 and blaNDM–5 and blaOXA–181 from the same patient. Front Microbiol. 2020;10:3095. doi:10.3389/fmicb.2019.03095
21. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing.
22. European Committee on Antimicrobial Susceptibility Testing. Breakpoint Tables for Interpretation of MICs and Zone Diameters. Version 12.0. Växjö, Sweden: European Committee on Antimicrobial Susceptibility Testing; 2022.
23. Rogers BA, Sidjabat HE, Silvey A, et al. Treatment options for New Delhi metallo-beta-lactamase-harboring Enterobacteriaceae. Microb Drug Resist. 2013;19(2):100–103. doi:10.1089/mdr.2012.0063
24. Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi:10.1038/s41426-018-0124-z
25. Nour I, Eldegla HE, Nasef N, Shouman B, Abdel-Hady H, Shabaan AE. Risk factors and clinical outcomes for carbapenem-resistant Gram-negative late-onset sepsis in a neonatal intensive care unit. J Hosp Infect. 2017;97(1):52–58. doi:10.1016/j.jhin.2017.05.025
26. Chiotos K, Hayes M, Gerber JS, Tamma PD. Treatment of carbapenem-resistant Enterobacteriaceae infections in Children. J Pediatric Infect Dis Soc. 2020;9(1):56–66. doi:10.1093/jpids/piz085
27. Montagnani C, Tersigni C, D’Arienzo S, et al. Resistance patterns from urine cultures in children aged 0 to 6 years: implications for empirical antibiotic choice. Infect Drug Resist. 2021;14:2341–2348. doi:10.2147/IDR.S293279
28. Li S, Chen Z, Huang L, et al. Safety of quinolones in children: a systematic review and meta-analysis. Paediatr Drugs. 2022;24(5):447–464. doi:10.1007/s40272-022-00513-2
29. Rzeczkowska M, Wołkowicz T, Zacharczuk K, et al. Draft genome sequence of an Escherichia coli ST410 isolate co-harbouring blaCTX-M-15, blaCMY-42, blaOXA-1, aac(3)-IIa and aac(6’)-Ib-cr genes with gyrA and parC mutations isolated from a paediatric patient in Poland. J Glob Antimicrob Resist. 2019;16:120–122. doi:10.1016/j.jgar.2018.11.024
30. Roer L, Overballe-Petersen S, Hansen F, et al. Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere. 2018;3(4):e00337–18. doi:10.1128/mSphere.00337-18
31. Piazza A, Comandatore F, Romeri F, et al. First report of an ST410 OXA-181 and CTX-M-15 coproducing Escherichia coli clone in Italy: a whole-genome sequence characterization. Microb Drug Resist. 2018;24(8):1207–1209. doi:10.1089/mdr.2017.0366
32. Kim JS, Yu JK, Jeon SJ, et al. Dissemination of an international high-risk clone of Escherichia coli ST410 co-producing NDM-5 and OXA-181 carbapenemases in Seoul, Republic of Korea. Int J Antimicrob Agents. 2021;58(6):106448. doi:10.1016/j.ijantimicag.2021.106448
33. Zhong Y, Guo S, Schlundt J, Kwa AL. Identification and genomic characterization of a blaNDM-5-harbouring MDR plasmid in a carbapenem-resistant Escherichia coli ST410 strain isolated from a natural water environmental source. JAC Antimicrob Resist. 2022;4(4):dlac071.
34. Devanga Ragupathi NK, Vasudevan K, Venkatesan M, Veeraraghavan B. First Indian report on B4/H24RxC ST410 multidrug-resistant Escherichia coli from bloodstream infection harbouring blaOXA-181 and blaCTX-M-15. J Glob Antimicrob Resist. 2020;22:568–570. doi:10.1016/j.jgar.2020.06.013
35. Gu J-N, Chen L, Weng X-B, Yang X-Y, Pan D-M. Clinical and microbiological characteristics of a community-acquired carbapenem-resistant Escherichia coli ST410 isolate harbouring blaNDM-5-encoding IncX3-type plasmid from blood. Front Med. 2021;8:658058. doi:10.3389/fmed.2021.658058
36. Negeri AA, Mamo H, Gurung JM, et al. Antimicrobial resistance profiling and molecular epidemiological analysis of extended spectrum β-lactamases produced by extraintestinal invasive Escherichia coli isolates from Ethiopia: the presence of international high-risk clones ST131 and ST410 revealed. Front Microbiol. 2021;12:706846. doi:10.3389/fmicb.2021.706846
37. Chen L, Peirano G, Kreiswirth BN, Devinney R, Pitout JDD. Acquisition of genomic elements were pivotal for the success of Escherichia coli ST410. J Antimicrob Chemother. 2022;77(12):3399–3407. doi:10.1093/jac/dkac329
38. Magaña-Lizárraga JA, Gómez-Gil B, Rendón-Maldonado JG, Delgado-Vargas F, Vega-López IF, Báez-Flores ME. Genomic profiling of antibiotic-resistant Escherichia coli isolates from surface water of agricultural drainage in North-Western Mexico: detection of the international high-risk lineages ST410 and ST617. Microorganisms. 2022;10(3):662.
39. Furlan J, Gonzalez I, Ramos PL, Stehling EG. International high-risk clone of multidrug-resistant CTX-M-8-producing Escherichia coli C-ST410 infecting an elephant (Loxodonta africana) in a zoo. J Glob Antimicrob Resist. 2020;22:643–645. doi:10.1016/j.jgar.2020.06.018
40. Ortega-Paredes D, Barba P, Mena-López S, Espinel N, Zurita J. Escherichia coli hyperepidemic clone ST410-A harboring blaCTX-M-15 isolated from fresh vegetables in a municipal market in Quito-Ecuador. Int J Food Microbiol. 2018;280:41–45. doi:10.1016/j.ijfoodmicro.2018.04.037
41. Wu W, Feng Y, Tang G, Qiao F, McNally A, Zong Z. NDM metallo-β-lactamases and their bacterial producers in health care settings. Clin Microbiol Rev. 2019;32(2):e00115–18. doi:10.1128/CMR.00115-18
42. Varani A, He S, Siguier P, Ross K, Chandler M. The IS6 family, a clinically important group of insertion sequences including IS26. Mob DNA. 2021;12(1):11. doi:10.1186/s13100-021-00239-x
43. Pong CH, Harmer CJ, Ataide SF, Hall RM. An IS 26 variant with enhanced activity. FEMS Microbiol Lett. 2019;366(3):fnz031. doi:10.1093/femsle/fnz031
44. Chowdhury G, Ramamurthy T, Das B, et al. Characterization of NDM-5 carbapenemase-encoding gene (blaNDM-5) – positive multidrug resistant commensal Escherichia coli from diarrheal patients. Infect Drug Resist. 2022;15:3631–3642. doi:10.2147/IDR.S364526
45. Zou H, Berglund B, Wang S, et al. Emergence of blaNDM-1, blaNDM-5, blaKPC-2 and blaIMP-4 carrying plasmids in Raoultella spp. in the environment. Environ Pollut. 2022;306:119437. doi:10.1016/j.envpol.2022.119437
46. Stokes HW, Tomaras C, Parsons Y, Hall RM. The partial 3′-conserved segment duplications in the integrons In6 from pSa and In7 from pDGO100 have a common origin. Plasmid. 1993;30(1):39–50. doi:10.1006/plas.1993.1032
47. Lallement C, Pasternak C, Ploy M-C, Jové T. The role of ISCR1-borne POUT promoters in the expression of antibiotic resistance genes. Front Microbiol. 2018;9:2579. doi:10.3389/fmicb.2018.02579
48. Nowrotek M, Kotlarska E, Łuczkiewicz A, Felis E, Sochacki A, Miksch K. The treatment of wastewater containing pharmaceuticals in microcosm constructed wetlands: the occurrence of integrons (int1–2) and associated resistance genes (sul1–3, qacEΔ1). Environ Sci Pollut Res Int. 2017;24(17):15055–15066. doi:10.1007/s11356-017-9079-1
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.