Back to Journals » Cancer Management and Research » Volume 10
The expression profile and clinical application potential of hsa_circ_0000711 in colorectal cancer
Authors Li J, Ni S, Zhou C , Ye M
Received 26 April 2018
Accepted for publication 11 June 2018
Published 17 August 2018 Volume 2018:10 Pages 2777—2784
DOI https://doi.org/10.2147/CMAR.S172388
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Kenan Onel
Jinyun Li,1 Shumin Ni,1 Chongchang Zhou,2 Meng Ye1
1Department of Oncology and Hematology, The Affiliated Hospital of Medical School of Ningbo University, Ningbo, China; 2Department of Otorhinolaryngology Head and Neck Surgery, Lihuili Hospital of Ningbo University, Ningbo, China
Introduction: Circular RNAs, as a class of long-time-ignored non-coding RNA, have been revealed as multifunctional RNAs in recent years, especially in the cancer research. However, the mechanism of most circular RNAs and their clinical application values in human cancers remain unknown, including in colorectal cancer (CRC).
Methods: In this study, we focused on the expression pattern and clinical values of hsa_circ_0000711 in CRC. The expression level of hsa_circ_0000711 in 101 paired CRC tissues and 3 CRC cell lines (HCT116, COLO205, and H-T29), as well as human normal colon epithelial cell line NCM460, were measured by quantitative real-time polymerase chain reaction.
Results: Our results revealed that the expression level of hsa_circ_0000711 was significantly downregulated in CRC tissues (P=9.35E-16) and CRC cell lines (P<0.01). In addition, the area under the receiver characteristic curves was 0.81. The sensitivity and specificity were 0.91 and 0.58, respectively. Meanwhile, our study showed that low expression of hsa_circ_0000711 could serve as an independent prediction biomarker associated with poor overall survival of CRC patients (hazard ratio =2.409; 95% CI: 1.276–4.547; P=0.004).
Conclusion: All of these results indicated that hsa_circ_0000711 may play a crucial role in CRC carcinogenesis and could be a potential effective biomarker for the diagnosis and prognosis of CRC.
Keywords: circular RNAs, biomarker, diagnosis, prognosis, CRC
Introduction
Colorectal cancer (CRC) is one of the most common gastrointestinal cancers and the fourth leading cause of cancer-related death in the world.1 The highest incidence of CRC is reported in countries of Europe, North America, and Oceania.2 According to epidemiologic data from the American Cancer Society, about 135,430 newly diagnosed and 502,600 deaths due to CRC were projected to occur only in the USA in 2017.3 Moreover, the rapidly increasing incidence of CRC in previously low-risk areas, such as Eastern Europe and East Asia, has been noted, which is attributed to the changes in dietary patterns and risk factors toward a so-called western lifestyle.4 Currently, the cornerstones of therapy for CRC are surgery, neoadjuvant radiotherapy, and adjuvant chemotherapy. However, the outcome of CRC remains unsatisfactory and the 5-year survival rate is <65%.5 Local recurrence and distant metastasis are the main causes leading to poor outcome of CRC.6 The 5-year survival rate of CRC dramatically declines from ~90% in early-stage non-metastatic tumors to about 5% in cases with distant metastasis.7 Therefore, precise prediction at the molecular level of relapse and identify effective early biomarkers for CRC are urgently needed for individual diagnosis and therapy.
Circular RNAs (circRNAs) are a novel class of non-coding RNAs (ncRNAs) and single-stranded covalently closed circular transcripts without 5′ caps and 3′ tails.8,9 In the beginning, circRNAs were regarded as splicing errors or by-products of RNA processing with low abundance.10 However, owing to the development of the next-generation RNA sequencing and bioinformatic analysis, researchers have discovered thousands of circRNAs and numerous studies have shed light on the biogenesis and function of circRNAs.11,12 Emerging evidence has showed that circRNAs are involved in the progression of tumorigenesis, invasion, and metastasis in numerous human carcinomas by regulating microRNA (miRNA) function as miRNA sponges.13 Additionally, compared with the linear RNAs, circRNAs expression is characterized as tissue-specific and stable which can not be degraded by RNase.14,15 Furthermore, numerous evidence supported that some circRNAs can stably exist in human body fluid.16 All these indications suggested that circRNAs could be served as a promising biomarker for cancer diagnosis and prognosis.17–19
Searching CRC-associated circRNAs by synthetically bioinformatic analysis in 2 circRNA databases: CircBase and circ2Traitsby,20,21 we focused on circRNA- hsa_circ_0000711 in this study. The position of hsa_circ_0000711 is located at chr16:68155889–68160513 with 4,624 nts of the spliced sequence length in gene symbol NFATC3. In this study, we first investigated the hsa_circ_0000711 expression characteristics in CRC tissues and corresponding normal tissues, as well as cell lines, and evaluated the association between the hsa_circ_0000711 expression level and clinicopathological characteristics of patients with CRC. Moreover, we also explored its diagnostic and prognostic potential for CRC.
Materials and methods
Patient characteristics and tissue specimen collection
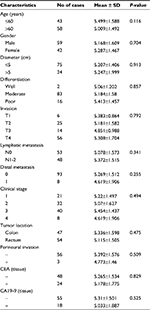
A total of 101 CRC tissues and paired adjacent nontumorous tissues were collected from CRC surgical specimens at the Department of gastrointestinal surgery in Ningbo Lihuili Hospital between June 2010 and April 2015. None of the patients underwent chemotherapy or radiotherapy before surgery. All the specimens were immediately preserved in RNA-fixer reagent after surgical excision and stored in liquid nitrogen. The final diagnosis was confirmed by 2 experienced pathologists. There were 2 well-differentiated, 83 moderately differentiated, and 16 poorly differentiated cases. Tumors were staged according to the TNM stage system of the American Joint Committee on Cancer seventh edition (2010). There were 21 Stage I, 32 Stage II, 40 Stage III, and 8 Stage IV cases. Clinicopathologic characteristics were available for each of the 101 patients. Patients were followed up for 60 months. Median follow-up was 39 months, with an interquartile range of 29–50 months. During follow-up, 5 patients were lost and 42 died. All patients agreed to participate in the study and signed the informed consent for the tissue collection. The study was approved by the Human Research Ethical Committee of Ningbo Lihuili Hospital.
Cell culture
Human normal colon epithelial cell line NCM460, as well as human CRC cell lines HCT116, COLO205, and HT29 were obtained from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Roswell Park Memorial Institute-1640 Medium (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (ExCell Biology, Shanghai, China) with 50 U/mL penicillin and 50 g/mL streptomycin (HyClone). The incubators were maintained at 37°C in a 5% CO2 atmosphere. Exponentially growing cells were used for experiments.
Total RNA extraction and quantitative real-time polymerase chain reaction (PCR) assay
Total RNA was first extracted from tissues and cells using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA), and then reversed transcription into cDNA by GoScript Reverse Transcription (RT) System (Promega Corporation, Fitchburg, WI, USA) following the manufacturer’s protocol. The divergent primers for amplified sequences of hsa_circ_0000711 were: forward, 5′-CACTAGACTGGCCTTTACC-3′ and reverse, 5′-CACAATCATCTGGCTCAA-3′. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was simultaneously amplified as internal control.22 Each reaction mixture (20 µL) contained 5 µL cDNA, 1 µL forward primer, 1 µL reverse primer, 10 µL SYBR Green I Master (Hoffman-La Roche Ltd., Basel, Switzerland), and 3 µL DNA/RNA-free water (Hoffman-La Roche Ltd.). The PCR amplification was performed in 384-well plates using the RocheLightCycler 480II instrument (Hoffman-La Roche Ltd.). The PCR reaction was conducted using the following conditions: 95°C for 10 minutes, amplification for 45 cycles at 95°C for 20 seconds, 60°C for 30 seconds, and 72°C for 40 seconds. A melting curve step was performed at 95°C for 15 seconds, 1 minute at 60°C, and then increasing temperature at 0.11°C per second for up to 95°C to measure fluorescence signal. The cycle threshold (Ct) values were recorded for both hsa_circ_0000711 and GAPDH. The expression level of hsa_circ_0000711 was calculated using the ΔCt method, higher ΔCt means lower expression. All the samples were assayed in triplicate.
Immunohistochemical analysis of tissue carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19-9)
We incubated the paraffin tissue sections in primary anti- CEA or anti-CA19-9 (Dako Denmark A/S, Glostrup, Denmark) for 1 hour at room temperature, and then, the tissues were incubated in diaminobenzidine (Dako Denmark A/S) for color development after incubation with broad-spectrum second antibody K5007 (Dako Denmark A/S). The standard for determination of the results was according to the 2010 American Society of Clinical Oncology/the College of American Pathologists guideline.
Statistical analyses
All statistical analyses were performed by using SPSS Version 18.0 (SPSS Inc., Chicago, IL, USA). Differences in continuous data between pairs of groups were detected by Student’s t-test, one-way analysis of variance test was applied between multiple groups (>2 groups). The diagnostic value of hsa_circ_0000711 was assessed using the receiver operating characteristic (ROC) curve and the area under the curve (AUC). Survival analyses were conducted by Kaplan–Meier and Cox proportional hazard regression analyses. A 2-tailed P-value of <0.05 was considered statistically significant. All the figures were drawn using the GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA).
Results
Expression of hsa_circ_0000711 in CRC tissues and cell lines
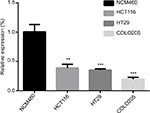
We explored hsa_circ_0000711 expression levels in 101 paired CRC tissues and corresponding normal tissues, the sequence of the qRT-PCR product was coincident with that in circBase (http://circbase.org/; Figure 1). The result showed that its expression in CRC tissues was significantly lower than that in adjacent normal tissues (P=9.35E-16, Figure 2). Among 101 paired CRC samples tested in our study, 76 cases (79.2%) exhibited significant downregulation of has_circ_0000711 in cancerous tissues (Figure 3). Then, we found that the expression levels of hsa_circ_0000711 in 3 CRC cell lines, HCT116, COLO205, and HT29, were significantly downregulated than those in normal colon epithelial cell line NCM460 (Figure 4).
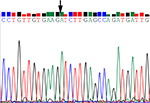  | Figure 1 DNA sequencing results of hsa_circ_0000711 in colorectal cancer tissues. Note: The arrow points to the cyclation site. |
  | Figure 2 hsa_circ_0000711 expression levels in 101 paired colorectal tissues and corresponding normal tissues. Abbreviation: Ct, cycle threshold. |
  | Figure 3 The expression level of hsa_circ_0000711 was significantly downregulated in 79.2% (76/96) colorectal cancer tissues compared with the adjacent normal tissues. |
Association between hsa_circ_0000711 expression levels and clinicopathological factors in CRC patients
Based on the aforementioned findings, we analyzed the association between hsa_circ_0000711 expression and clinicopathological characteristics of patients with CRC, such as age, gender, diameter, differentiation, invasion, lymph node metastasis, clinical stage, distant metastases, location, perineural invasion, and CEA and CA19-9 levels. However, no difference of hsa_circ_0000711 expression was found among these clinicopathological characteristics of CRC patients (Table 1).
The diagnostic value of hsa_circ_0000711 in CRC
The ROC curve was used to evaluate the diagnostic potential of hsa_circ_0000711 in CRC patients. The ROC curve is a comprehensive index that can reflect on the specificity and sensitivity of continuous variables. The maximum Youden index was used as a cutoff point. In current study, Figure 5 showed the AUC was up to 0.81 at a cutoff ΔCt value of 3.37. The sensitivity and specificity were 0.91 and 0.58, respectively. Hence, hsa_circ_0000711 might be a diagnostic indicator of CRC.
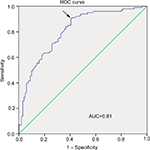  | Figure 5 ROC curve. Abbreviations: AUC, area under the curve; ROC, receiver operating characteristic. |
The prognostic value of hsa_circ_0000711 in CRC
We constructed survival curves to investigate the prognostic value of hsa_circ_0000711 for CRC (Figure 6). The median expression was used as a cutoff point. A total of 51 patients were assigned to the low-expression group, and the others to the high-expression group. The Kaplan–Meier survival analysis revealed that low-expression level of hsa_circ_0000711 was significantly associated with poor outcome of CRC (log-rank P=0.003). In univariate Cox proportional hazards analysis, patients with low expression had a significantly increased risk of death compared with patients with high expression of hsa_circ_0000711 (hazard ratio [HR]=2.418; 95% CI: 1.315–4.446; P=0.005). Subsequently, a multivariate Cox proportional hazard analysis was performed by adjusting for T classification and lymphatic metastasis. The results indicated low expression of hsa_circ_0000711 (HR=2.409; 95% CI: 1.276–4.547; P=0.004) and lymphatic metastasis (HR=2.045; 95% CI: 1.105–3.785; P=0.023) was an independent predictive biomarker of overall survival (OS) of CRC patients (Table 2).
  | Figure 6 The Kaplan–Meier survival curve. Note: The median ΔCt value was used as a cutoff point. Abbreviation: Ct, cycle threshold. |
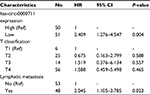  | Table 2 Multivariate Cox proportional hazards analysis of the 101 colorectal cancer patients Abbreviations: Ref, reference category; HR, hazard ratio. |
Discussion
Owing to advancements in high-throughput sequencing technologies and bioinformatics, circRNAs, once perceived as the by-products of splicing errors, recently were found to be extensively expressed in human cells and play roles in many biological processes.15,23 Although, circRNAs are commonly grouped into a type of ncRNA, recent study showed that circRNAs can be translated into proteins,24,25 which enriched circRNAs’ critical roles in life activity. In addition, mounting evidence shows that circRNAs are widely involved in the initiation and progression of multiple diseases, especially cancers.26 For example, Deng et al disclosed that hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma.27 Yao et al demonstrated that circZKSCAN1 inhibits growth, migration, and invasion of hepatocellular carcinoma via several cancer-related signaling pathways.28 Due to lack of the 5′cap and polyA tail, circRNAs are highly conserved and remarkably stable.29,30 In addition, circRNAs are generally expressed in a tissue- and cell-type-specific manner and could help distinguish different tumors or predict treatment responses.31 Taken together, these characteristics exhibit that circRNAs could be potential to serve as a kind of valuable biomarker of cancer diagnosis and prognosis.
Up to now, increasing researches and achievements reported that circRNAs have involved in CRC progression. Xie et al revealed that hsa_circ_001569 was a positive regulator in cell proliferation and invasion of CRC by as a sponge of miR-145.32 Wang et al found that hsa_circ_001988 was decreased expression and correlated with differentiation and perineural invasion of CRC.33 In the current study, our findings first showed that hsa_circ_0000711 expression level was significantly downregulated in both CRC tissues and CRC cell lines. Although previous studies showed that circRNAs might be correlated with clinicopathological parameters, no difference in hsa_circ_0000711 expression levels were found for clinicopathological characteristics of CRC patients in our study. Future rigorous clinical research studies with larger sample sizes will be essential to validate our findings.
Diagnosis of CRC patients depends on clinical symptoms and auxiliary examinations, such as colonscopy, CT colonography, MRI, and PET/CT colonography.34 However, due to non-specific symptoms in the early stage of CRC, and lack of effective diagnostic markers with high sensitivity and specificity, a low early diagnostic rate poses challenges for treatment. Because of the lack of free ends, circRNAs are resistant to exonucleases and more stable than linear RNA isoforms.15 Besides, targeted validation of the accuracy of a circRNA is accomplished by PCR using outward-facing primers and Sanger sequencing for a semi-random primer, and the technology is well developed.35 Due to their special structure, circRNAs can act as effective biomarkers for the early diagnosis of cancer.36–38 In this study, we also explored the value of hsa_circ_0000711 in CRC diagnosis by ROC curve, which is a comprehensive index used to reflect the sensitivity and specificity of continuous variables. The larger the area under the ROC curve (AUC) is, the higher diagnostic value is.39 In our study cohort, we found that the AUC was 0.81 and the sensitivity was 0.91. Compared with the diagnostic value of conventional CRC-related biomarkers, such as CEA and CA199,40 hsa_circ_0000711 had a higher AUC value. Meanwhile, we noticed that the specificity of hsa_circ_0000711 was not so high, suggesting that hsa_circ_0000711 cannot be used to diagnose CRC independently. However, given its near-perfect sensitivity, hsa_circ_0000711 might be an effective biomarker for the diagnosis of CRC if combined with other diagnostic technologies. Due to a relatively small sample size, the rigorous clinical research studies with larger sample sizes will be essential to corroborate our finding.
Although classification according to TNM and Union for International Cancer Control stage provides valuable prognostic information and guides therapy decisions, the response and outcome of individual patient’s therapy is not predicted enough.41 Increasing evidence showed that the circRNAs could be prognostic markers for cancers, including CRC.42 In our study, log-rank test showed that low expression level of hsa_circ_0000711 was significantly associated with poor OS of CRC patients, which was in accordance with our univariate Cox proportional hazards analysis results. In addition, a multivariate Cox proportional hazard analysis was performed by adjusting for T classification and lymphatic metastasis to confirm that low expression level of hsa_circ_0000711 was an independent unfavorable factor for CRC outcomes. All these findings supported the fact that hsa_circ_0000711 expression level could be a potential biomarker for prognosis of CRC and guide the individualized treatment programs.
Conclusion
To sum up, the present study indicates that circRNA0003906 was significantly downregulated in CRC and may play a crucial role in CRC carcinogenesis. Additionally, our finding supported that hsa_circ_0005986 might be a potential effective biomarker for the diagnosis and prognosis of CRC.
Acknowledgements
This work was supported by the Zhejiang Provincial Natural Science Foundation of China (grant number LY16H160005); the Project of Scientific Innovation Team of Ningbo (grant number 2015B11050); the Ningbo Natural Science Foundation (grant number 2014A610235 and 2017A610236).
Disclosure
The authors report no conflicts of interest in this work.
References
Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014; 383(9927):1490–1502. | ||
Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1688–1694. | ||
Miller KD, Siegel RL, Lin CC, et al. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–289. | ||
Zippi M, de Toma G, Minervini G, et al. Desmoplasia influenced recurrence of disease and mortality in stage III colorectal cancer within five years after surgery and adjuvant therapy. Saudi J Gastroenterol. 2017;23(1):39–44. | ||
Pizzini S, Bisognin A, Mandruzzato S, et al. Impact of microRNAs on regulatory networks and pathways in human colorectal carcinogenesis and development of metastasis. BMC Genomics. 2013;14:589. | ||
Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. | ||
Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859(1):163–168. | ||
Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature. 1986;323(6088):558–560. | ||
Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32(5):453–461. | ||
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. | ||
Xia W, Qiu M, Chen R, et al. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. | ||
Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143(11):1838–1847. | ||
Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. | ||
Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. | ||
Sun H, Tang W, Rong D, et al. Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomark. 2018;21(2):299–306. | ||
Yang F, Liu DY, Guo JT, et al. Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol. 2017;23(47):8345–8354. | ||
Zhu X, Wang X, Wei S, et al. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284(14):2170–2182. | ||
Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20(11):1666–1670. | ||
Ghosal S, das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet. 2013;4:283. | ||
Shen Z, Li Q, Deng H, Lu D, Song H, Guo J. Long non-coding RNA profiling in laryngeal squamous cell carcinoma and its clinical significance: potential biomarkers for LSCC. PLoS One. 2014;9(9):e108237. | ||
Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. | ||
Legnini I, di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66(1):22–37. | ||
Pamudurti NR, Bartok O, Jens M. Translation of circRNAs. Mol Cell. 2017;66(1):9–21.e7. | ||
Qu S, Zhong Y, Shang R, et al. The emerging landscape of circular RNA in life processes. RNA Biol. 2017;14(8):992–999. | ||
Deng N, Li L, Gao J, et al. Hsa_circ_0009910 promotes carcinogenesis by promoting the expression of miR-449a target IL6R in osteosarcoma. Biochem Biophys Res Commun. 2018;495(1):189–196. | ||
Yao Z, Luo J, Hu K, et al. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. | ||
Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. | ||
Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. | ||
Qu S, Liu Z, Yang X, et al. The emerging functions and roles of circular RNAs in cancer. Cancer Lett. 2018;414:301–309. | ||
Xie H, Ren X, Xin S, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7(18):26680–26691. | ||
Wang X, Zhang Y, Huang L, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol. 2015;8(12):16020–16025. | ||
Kijima S, Sasaki T, Nagata K, Utano K, Lefor AT, Sugimoto H. Preoperative evaluation of colorectal cancer using CT colonography, MRI, and PET/CT. World J Gastroenterol. 2014;20(45):16964–16975. | ||
Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. | ||
Chen S, Li T, Zhao Q, Xiao B, Guo J. Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta. 2017;466:167–171. | ||
Qin M, Liu G, Huo X, et al. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16(1):161–169. | ||
Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379. | ||
Jones CM, Athanasiou T. Summary receiver operating characteristic curve analysis techniques in the evaluation of diagnostic tests. Ann Thorac Surg. 2005;79(1):16–20. | ||
Feng B, Zheng MH, Zheng YF, et al. Normal and modified urinary nucleosides represent novel biomarkers for colorectal cancer diagnosis and surgery monitoring. J Gastroenterol Hepatol. 2005;20(12):1913–1919. | ||
Kong X, Li J, Cai Y, et al. A modified TNM staging system for non-metastatic colorectal cancer based on nomogram analysis of SEER database. BMC Cancer. 2018;18(1):50. | ||
Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.