Back to Journals » OncoTargets and Therapy » Volume 13
The Expression and Prognostic Significance of Claudin-8 and Androgen Receptor in Breast Cancer
Authors Zhang Y, Zheng A, Lu H, Jin Z, Peng Z, Jin F
Received 14 December 2019
Accepted for publication 3 April 2020
Published 23 April 2020 Volume 2020:13 Pages 3437—3448
DOI https://doi.org/10.2147/OTT.S242406
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Yiqi Zhang,1 Ang Zheng,1 Heng Lu,2 Zining Jin,1 Ziqi Peng,1 Feng Jin1
1Department of Breast Surgery, The First Affiliated Hospital of China Medical University, Shenyang, People’s Republic of China; 2Department of Thoracic Surgery, The First Affiliated Hospital of Jinzhou Medical University, Jinzhou, People’s Republic of China
Correspondence: Feng Jin
Department of Breast Surgery, The First Affiliated Hospital of China Medical University, Shenyang City, Liaoning Province 110004, People’s Republic of China
Tel +86 18841617682
Email [email protected]
Purpose: Claudin-8 (CLDN8) has been identified as an androgen-regulated gene in prostate cancer. However, the role of CLDN8 has not been fully explored in breast cancer. We aimed to explore the expression of CLDN8 and androgen receptor (AR), determine the correlation between CLDN8 and AR, assess the prognostic value of CLDN8 and AR co-expression, and investigate the possible CLDN8 expression molecular mechanism in breast cancer.
Materials and Methods: Twenty-eight pairs of fresh tumor tissues and adjacent noncancerous tissues were evaluated by Western blot for CLDN8. Then, 142 breast cancer samples were determined by immunohistochemistry for CLDN8 and AR. The association of clinicopathological features with CLDN8, AR and CLDN8, and AR co-expression was examined. The Cancer Genome Atlas (TCGA) was used to demonstrate the expression of CLDN8 and correlation between CLDN8 and AR. Kaplan–Meier survival analysis was performed to assess the prognostic impact of CLDN8 and AR co-expression. The mechanisms related to CLDN8 expression in breast cancer were explored by Gene Set Enrichment Analysis (GSEA).
Results: CLDN8 was downregulated in breast cancer tissues and positively correlated with none lymph node metastasis (P=0.016), low histological grade (P=0.006), positive ER (P=0.014), positive PR (P=0.003), low Ki-67 index (P=0.017) and molecular subtypes (P=0.012). CLDN8 level was significantly associated with AR level (r=0.348; P< 0.001). CLDN8 and AR co-expression was positively correlated with none lymph node metastasis (P=0.007), low histological grade (P=0.017), positive ER (P=0.019), positive PR (P=0.015) and low Ki-67 index group (P=0.038). CLDN8 and AR co-expression had a better clinical prognosis.
Conclusion: The expression of CLDN8 is directly related to the expression of AR. CLDN8 and AR co-expression might be a potential prognostic evaluation factor for breast cancer patients.
Keywords: claudin-8, androgen receptor, breast cancer, prognosis
Introduction
The heterogeneity and the distinction among different molecular subtypes of breast cancer require a further understanding of predictive and prognostic factors to improve their survival. Like estrogen receptor (ER) and progesterone receptor (PR), androgen receptor (AR) is a member of steroid nuclear receptor family.1 Although androgen and AR are generally considered extremely important in the physiological and pathological process of prostate tissue, the majority of breast cancers also express AR, regardless whether ER is positive or not, and their effect in breast cancer prognosis and therapy has long been highlighted.2 Functionally, AR expression has a dual regulatory effect on breast cancer. AR expression is highly concordant to ER expression and involved in a complex cross-talk with ER.3 In ER-positive (ER+) and AR-positive (AR+) breast cancer, AR leads to cell apoptosis and act as a favorable prognostic factor by binding estrogen-responsive element.4,5 But in ER-negative (ER-) and AR+ breast cancer (termed molecular apocrine phenotype), AR leads to cell proliferation and act as a poor prognostic factor by binding androgen-responsive element.6 Indeed, these data suggest that evaluation AR status may offer additional information for breast cancer patients in terms of prognosis and provide opportunities for AR targeted therapy.
Claudins, together with occludins and junctional adhesion molecules, form a fundamental structure of tight junctions by cell–cell adhesions to modulate epithelial paracellular transport.7 Until now, at least 27 different members of the claudin family varying from 22 to 27 kD have been identified.8 Dysregulation of claudin expression leads to tight junction loss function and then promotes tumorigenesis.9 Interestingly, previous analyses have reported that claudin-low breast cancer which is largely within triple-negative breast cancer (TNBC) confers high malignancy and poor survival.10,11
Reduced level of claudin-8 (CLDN8), which is caused by androgen ablation, is associated with inflammatory infiltrate in mice prostate.12 AR is involved in CLDN8 transcriptional activation in prostate cancer.13 Ashikari et al reported consistently that AR regulates CLDN8 expression and facilitate the development of prostate cancer.14 CLDN8 has been found primarily expressed in breast cell lines in a cell-specific manner.15 CLDN8 was clustered in ER-positive subgroup and was proposed as “luminal” claudin among high-grade breast invasive ductal carcinoma.16 RNA sequencing analysis showed that CLDN8 is a gene targeted by AR and markedly increased in the presence of DHT.17 However, the expression of CLDN8 and the correlation between CLDN8 and AR in breast cancer patients have not been well elucidated.
In this study, we used The Cancer Genome Atlas-breast cancer (TCGA-BRCA) to obtain the expression level of CLDN8 and AR at mRNA level. After that, we investigated the expression of the CLDN8 and AR at protein level and identified their relationship with various clinicopathological parameters in breast cancer tissues. We determined their function on predicting the prognosis in breast cancer patients. Finally, we used Gene Set Enrichment Analysis (GSEA) to investigate CLDN8-related signaling pathways.
Materials and Methods
TCGA and Gene Set Enrichment Analysis (GSEA)
Gene expression (https://cancergenome.nih.gov/) from the TCGA-BRCA database was downloaded. It contains 113 normal breast samples and 1109 breast cancer samples. Then, we used the edgR package to normalize gene expression. The difference of CLDN8 expression in normal tissues and breast cancer tissues and the relationship between CLDN8 and AR were analyzed by GraphPad Prism 7.0. GSEA (http://www.broadinstitute.org/gsea/index.jsp) was analyzed to explore the CLDN8 expression mechanisms in breast cancer.
Patients and Tissue Samples
After the approval of the Research Ethics Committee of the First Affiliated Hospital of China Medical University, fresh tumor tissues and adjacent noncancerous tissues were obtained from 28 patients who were pathologically diagnosed with infiltrative breast carcinoma and underwent modified radical mastectomy at the First Affiliated Hospital of China Medical University. Every patient signed an informed consent form. All the samples were frozen at −80°C within 30 minutes after surgical resections. Protein was extracted from these samples to perform Western blot analysis. Another 142 primary non-metastatic breast cancer specimens were retrospectively collected from the above hospital between 2011 and 2013 to perform immunohistochemical staining. No patients had received any treatment before surgery, and patients who lost reliable medical records were excluded. While all the patients in this study received radical mastectomy, they did not undergo breast-conserving surgery. Based on the immunohistochemical expression of ER, PR, HER2 and Ki-67, the selected cases comprised 38 luminal A, 47 luminal B HER2-negative, 25 luminal B HER2-positive, 18 HER2-enriched and 14 triple-negative breast cancer. Every patient received the standard postmastectomy treatment based on the National Comprehensive Cancer Network (NCCN) Guidelines. Of the 142 cases, 26 received hormonal therapy alone, 14 received chemotherapy alone, 59 received both hormonal therapy and chemotherapy, 18 received chemotherapy and anti-HER2 therapy, 25 received hormone therapy chemotherapy and anti-HER2 therapy. Of the above cases, 83 received radiotherapy at the discretion of the treating radiation oncologist.
Data including age, tumor size, lymph node metastasis, histological grade, the status of ER, PR, HER2 and Ki-67 were obtained from the Hospital Information System and pathology diagnosis reports. We called patients and checked their medical records to collect their follow-up data.
Western Blotting
Total protein was extracted from the frozen tissues using lysis buffer on ice. Bradford method was used to quantify the protein. Equal amounts of protein (20µg) were subjected to 12% polyacrylamide, 0.1% SDS (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes (Millipore Corporation, Billerica, USA). Following blocking in phosphate-buffered saline with 0.1% Tween-20 (PBST) containing 0.5% skim milk powder for 1 hour at room temperature (RT), the membranes were incubated in different primary antibodies overnight at 4°C. The primary antibodies used were rabbit anti-CLDN8 antibody (1:10,000; Abcam, Cambridge, MA, USA) and mouse anti-GAPDH antibody (1:5000, Santa Cruz Biotechnology, CA, USA). After washing by PBST, the membranes were incubated with HRP-linked anti-rabbit or HRP-linked anti-mouse secondary antibodies for 1 hour at RT. The detection was carried out with an ECL Western blotting kit (BioVision, San Francisco, USA), the density of protein bands was analyzed by Adobe Illustrator CS6 analysis software.
Immunohistochemical Staining
Paraffin-embedded breast tissues were sliced into 4μm sections. These sections were deparaffinized and rehydrated in xylene and a series of graded alcohols. After immersed in 3% hydrogen peroxide at RT for 15 minutes, these sections had retrieved antigen in citrate buffer (pH 6.0) at 121°C for 90s. They were then incubated with primary antibody against CLDN8 (1:100; Abcam, Cambridge, MA, USA) and AR (1:100; Proteintech, Wuhan, China) overnight at 4°C. Phosphate-buffered saline (PBS) without primary antibodies was used as negative control. After washing, slides were incubated with biotinylated goat anti-rabbit/mouse immunoglobulin, followed by incubated with streptomycin avidin-peroxidase, basing on Ultra-sensitive™ S-P kit protocol (Maixin-Bio, Fuzhou, China). The specific immunostaining was then visualized with DAB reagent. The images were taken by an Olympus microscope.
Evaluation of Immunostaining
There were two blinded professional pathologists separately evaluated the slides. Each slide was evaluated from five visual fields and each field contains at least 100 cells at 200× magnification. The immunoreactivity of CLDN8 in breast tissues was categorized based on staining intensity scored as ‘0ʹ (negative), ‘1ʹ (weak), ‘2ʹ (moderate), or ‘3ʹ (strong) plus positive percentage scored as ‘0ʹ (none), ‘1ʹ (<1/100), ‘2ʹ (1/100 to 1/10), ‘3ʹ (1/10 to 1/3), ‘4ʹ (1/3 to 2/3), or ‘5ʹ (>2/3). The immunohistochemistry (IHC) scores were classified into two groups as follows: high expression (7–8) or low expression (0–6).14 The immunoreactivity of AR was assessed based on the positive percentage of nuclear staining cells: positive (≥10%) or negative (<10%).
Statistical Analysis
SPSS 23.0 (IBM, Armonk, USA) and GraphPad 7.0 (GraphPad Software, San Diego, USA) were used for statistical analyses. The chi-square tests were performed to analyze the association between protein expression and clinicopathological characteristics. The correlation between CLDN8 and AR was examined by Spearman correlation analysis. Cox regression was performed to analyze Overall survival (OS) and Disease-free survival (DFS). The survival differences between the CLDN8 and AR co-expression patients and other expression patients were assessed by Kaplan–Meier method. A p<0.05 was considered statistically significant.
Results
Expression of CLDN8 and AR in Breast Cancer
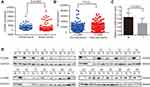
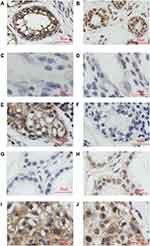
To investigate the role of CLDN8 in breast cancer, we first determined CLDN8 expression in TCGA datasets. The data proved that compared with normal tissues, the level of CLDN8 was significantly lower in tumor tissues (P<0.0001, Figure 1A). We evaluated CLDN8 expression levels in 28 fresh paired breast cancer tissues and adjacent noncancerous breast tissues by Western blot analysis. In accordance with our previous finding, the normalized expression levels of CLDN8 in breast cancer tissues were significantly lower compared to matched non-cancerous tissues (P<0.001, Figure 1C, D). Next, we evaluated the expression levels of CLDN8 and AR by IHC in 142 breast cancer samples and 40 adjacent normal breast tissues. The CLDN8 was mainly expressed in membranes (Figure 2A, E and I), while the AR was mainly expressed in nucleus (Figure 2B, H and J). CLDN8-positive expressed in 66/142 (46.5%) breast cancer cases, which was significantly lower than 26/40 (65.0%) in adjacent normal breast tissues (P<0.05). AR-positive expressed in 86/142 (60.6%) breast cancer cases, which was significantly lower than 31/40 (77.5%) in adjacent normal breast tissues (P<0.05). These results suggest that CLDN8 and AR low expression may be crucial for the breast tumorigenesis.
The Association of CLDN8 Expression and AR Expression with Clinicopathology Factors
The clinicopathological characteristics of the whole population and their association with CLDN8 expression and AR expression are shown in Table 1. The median follow-up time was 74.9 months (38–80) and the median age at diagnosis was 52.5 years (27–72). Age, tumor size and HER2 expression were not significantly associated with CLDN8 expression. However, we found CLDN8 was significantly higher expressed in patients with none lymph node metastasis (P=0.016), low histological grade (P=0.006), positive ER (P=0.014), positive PR (P=0.003) and low Ki-67 index (P=0.017), respectively. CLDN8 expression was significantly associated with molecular subtype (P=0.012). AR was significantly higher expressed in patients with positive ER (P=0.004) and low Ki-67 index (P=0.040). Moreover, we found a strict correlation between CLDN8 and AR expression (r=0.348, P<0.001, Table 2). The TCGA data also showed CLDN8 mRNA expression was positively correlated to AR mRNA expression (P<0.01, Figure 1B). These data indicate that in the breast cancer patients, CLDN8 and AR expression are positively related.
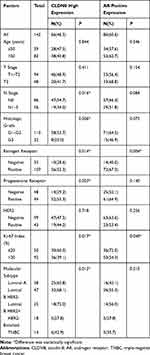 |
Table 1 The Relationship Between CLDN8 and AR Expression with Clinicopathological Features in Breast Cancer Patients |
 |
Table 2 Correlative Analysis of CLDN8 Expression with AR Expression |
There are four patterns of CLDN8 and AR expression, including CLDN8 negative and AR negative (Figure 2C and D), CLDN8 positive and AR negative (Figure 2E and F), CLDN8 negative and AR positive (Figure 2G and H), CLDN8 positive and AR positive (Figure 2I and J). As demonstrated in Table 3, in the whole patients, 36.7% of the cases showed CLDN8 and AR co-expression (CLDN8+/AR+). Compared with the other expression subgroup, there is a higher number of patients with CLDN8+/AR+ in the N0 (P=0.007), G1-2 (P=0.017), positive ER (P=0.019), positive PR (P=0.015) and low Ki-67 index group (P=0.038).
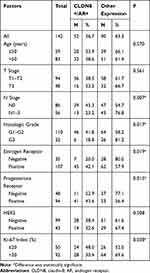 |
Table 3 The Relationship Between CLDN8/AR Co-Expression with Clinicopathological Features in Breast Cancer Patients |
The Impact of CLDN8/AR Co-Expression on Prognosis
In the Kaplan–Meier analysis, the OS rate of the CLDN8-positive group was significantly higher than the CLDN8-negative group (P=0.013, Figure 3A), while the DFS rate between the two groups had no statistical difference (P=0.088, Figure 3B). Interestingly, the DFS rate of AR-positive expression was obviously higher than the negative expression (P=0.043, Figure 3C), the OS rate between the two groups had no statistical difference (P=0.081, Figure 3D). Finally, we comprehensively decipher the prognosis impact of CLDN8/AR co-expression. As shown in Figure 4A and B, the CLDN8+/AR+ subgroup has longer OS and DFS compared with other expression subgroup (OS, P=0.005; DFS, P=0.034). We investigated the impact of CLDN8/AR co-expression on prognosis stratified for ER expression. The co-expression of CLDN8 and AR was associated with a longer OS compared to other expression subgroup in the ER-positive patients (P=0.032, Figure 4C), but may not provide more information on DFS (Figure 4D). We were unable to perform any analyses in the ER-negative patients since this phenotype only have 35 patients. These findings imply that lack of expression of CLDN8 and AR is associated with poor prognosis.
Then, we performed univariate and multivariate analyses to assess the impact of each clinicopathologic variable on prognosis in breast cancer patients. The association of OS and DFS with each clinicopathologic variable is summarized in Table 4. CLDN8+/AR+ was a significant factor associated with favorable OS (P=0.012) and DFS (P=0.039). Tumor size (OS, P=0.003; DFS, P<0.001), lymph node metastases (OS, P=0.001; DFS, P<0.001), HER2 (OS, P=0.019) were significant factors associated with shorter OS and DFS, respectively. Furthermore, multivariate analysis showed CLDN8+/AR+ (P=0.045) remained was an independent favorable prognostic factor for OS (Table 5). Tumor size (P=0.036), lymph node metastases (P=0.043) and HER2 (P=0.018) remained were poor prognostic factors for OS. Tumor size (P=0.005) and lymph node metastases (P=0.001) remained were poor prognostic factors for DFS.
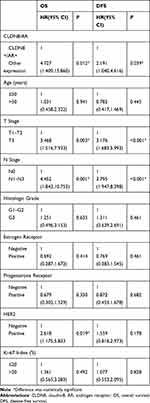 |
Table 4 Univariate Analysis of OS and DFS for Breast Cancer Patients |
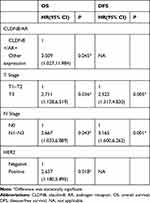 |
Table 5 Multivariate Analysis of OS and DFS for Breast Cancer Patients |
CLDN8-Related Signaling Pathways
To determine the identity of CLDN8-related biological pathway, we chose significant enrichment of genes basing on Normalized Enrichment Score (NES) (Figure 5A–H and Table 6). The result indicated that CLDN8 expression was enriched in pathways of androgen response, epithelial to mesenchymal transition (EMT), Wnt signaling pathway, Notch signaling, P53 pathway, pathways in cancer, TNFα signaling, JAK-STAT signaling pathway, etc.
 |
Table 6 Gene Set Enriched with CLDN8 High Expression |
Discussion
It is clear that as a member of tight junctions, CLDN8 is required for a paracellular barrier. Decreasing expression of CLDN8 leads to a collapse of the tight junction barrier and might have an extreme influence on cancer cells' survival and proliferation.12 The TCGA database analysis showed that the CLDN8 mRNA level was downregulated in breast cancer tissue. Consisting of mRNA expression level, Western blotting and IHC results also indicated that CLDN8 expression was significantly lower at protein level. Low CLDN8 expression was associated with lymph nodes metastasis, high histological grade, negative ER, negative PR, high Ki-67 index. As to various molecular subtyping groups, CLDN8 expression was significantly high in luminal breast cancer, but there was no significant difference among luminal subtypes. The claudin-low intrinsic subtype of breast cancer is identified by negative for three other members of the CLDN family, including CLDN3, 4 and 7. Tumor initiating cell features are enriched and common epithelial cell features are lacking in claudin-low tumors. Like CLDN8 low expression in our study, the claudin-low phenotype is also a poor prognosis factor of breast cancer.18
Hormonal therapy targeting the ER pathway has been intensively studied and well understood. However, a major obstacle is the inevitable drug resistance, continuing efforts to use new targeting drugs are needed. To date, as a new molecular target of breast cancer, AR has spurred increased attention directly. The successful experience in prostate cancer therapy gives confidence in breast cancer treatment. Approximately 60% to 80% of breast cancer are AR positive, depending on antibodies and definition of AR positivity.2,19,20 However, the role of androgenic actions in breast cancer remains controversial. Hence, this fuels the urgency to determine the role of AR in breast carcinomas. In our analysis, we found that AR was expressed in 60.6% of breast cancer and correlated to ER expression. Furthermore, AR was negatively related to the Ki-67 index, which is a predictor of proliferation and worse prognosis. It was reported that AR is significantly associated with positive prognostic factors.21 Luminal subtypes inclined to contain more tumors that were AR positive but there was no significant difference among molecular subtypes. The difference among luminal subtypes was not statistically significant. Surprisingly, AR and ER can utilize similar cis-regulatory regions, then AR can physically impair ER transcriptional activity.4 Several studies have demonstrated that co-expression of AR and ER predicts a beneficial clinical outcome.22,23 Additionally, AR targeted treatments in both preclinical and clinical findings for breast cancer are in development and have shown promising preliminary efficacy.24
The gene expression studies identified that CLDN8 is driven by AR in the luminal androgen receptor (LAR) tumors, which is a subtype of TNBC.25 It was reported that CLDN8 is an AR target gene and its transcriptional activation is up-regulated followed by DHT treatment in the breast cancer cells.17 We found CLDN8 mRNA level was co-related with AR mRNA level in the TCGA database analysis. To clarify the relation between CLDN8 and AR, we evaluated the expression status of CLDN8 and AR and identified a positive correlation between these two proteins. CLDN8 alone was correlated with better OS and AR alone was correlated with better DFS. Notably, patients with combined expression pattern of CLDN8 and AR correlated with better OS and DFS, which meant that the combined detection of CLDN8 and AR expression could provide a better comprehensive diagnosis for breast cancer patients. In the ER+ subgroup, co-expression of CLDN8 and AR had a better OS, but cannot bring more information on DFS. Taken together, a growing number of evidences support the fact that CLDN8 acts as a tumor-suppressing gene in breast cancer. In addition, CLDN8 is regulated by AR, co-expression of these two proteins strongly predicts favorable prognosis. Tumor size, lymph node metastases and HER2 expression were independent prognostic factors. However, such data still need to be well elucidated further in larger scale and more detailed investigations.
A multitude of signal pathways takes effect comprehensively during the multistep occurrence and development of tumors.26 GSEA provides meaningful insights in them.27 Several functional gene sets potentially correlated with CLDN8 expression were identified by GSEA in this study. In line with the previous study, CLDN8 might exert a response to androgen. Similar findings further support the conclusion that CLDN8 and AR expression is co-related. CLDN8 might contribute to EMT, in which the ability of a cell to invade and metastasize is increased. In this regard, a growing body of data demonstrates the crucial role of claudins in regulating the EMT process. Earlier studies had documented that claudin-low breast tumors are highly enriched for EMT transition markers.18 Similarly, the low expression of E-cadherin and high levels of vimentin in metaplastic breast carcinoma with low expression of claudin-3, -4, and -7 are two markers consistent with EMT features.10 Moreover, a similar parallel association of claudin-1 expression with EMT in colorectal cancer is well documented. Inhibiting claudin-1 expression in colorectal cancer cells not only leads to complex morphological changes but also result in EMT markers and associated functional behaviors change.28 As a published study has shown that CLDN family members are controlled by Wnt signal pathway, CLDN8 might also induce invasive and metastasis behavior of breast cancer via this pathway.29 In addition, CLDN8 might modulate barrier properties via Notch signaling. Interestingly, claudin-1 modulates this pathway in MMP-9 dependent manner in colon cancer.30 We also observed significant enrichment of genes relevant to P53 signaling, which indicated CLDN8 might participate in cellular growth and division. P53 cooperates with prolactin to promote proliferation, and aggressive behavior has been observed in claudin-low mammary carcinomas.31 Comprised CLDN8 variably was correlated to pathways of cancer implies that it may enhance properties of cancer cells such as tumorigenic, invasive, and metastatic. Furthermore, CLDN8 might causally involve in malignant phenotypes via the TNF-α signal pathway and JAK/STAT signal pathway. Taken together, CLDN8 seems to constitute an axis-of-evil in breast cancer, possibly by altering complex mechanisms at multiple levels. The exact regulations of CLDN8 in breast cancer remain to be fully clarified, and the efforts over the course of CLDN8 studies may help lead to new therapeutic targets.
Conclusion
In conclusion, the data presented here showed that the expression of CLDN8 was significantly low and CLDN8 expression level was correlated with AR expression level in breast cancer. In light of our results, the combination of CLDN8 and AR expression in breast cancer prognosis was defined as a strong favorable signature and should be further investigated as a potential biomarker for a new opportunity for therapy. Identifying pathways related to CLDN8 in breast cancer progression has the potential value to understand the molecular mechanism and may advance the treatment of breast cancer in future.
Ethics and Consent Statement
The study was approved by the Ethics Committee of China medical University. All procedures performed in this study were in accordance with the 1964 Helsinki Declaration and its later amendments.
Informed Consent
Written informed consent was obtained from all patients included in the study.
Acknowledgments
These studies were supported by the National Science Foundation of China (Grant No. 81773163). The authors would like to thank teachers and students for their help.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Tan MH, Li J, Xu HE, Melcher K, Yong EL. Androgen receptor: structure, role in prostate cancer and drug discovery. Acta Pharmacol Sin. 2015;36(1):3–23. doi:10.1038/aps.2014.18
2. Hu R, Dawood S, Holmes MD, et al. Androgen receptor expression and breast cancer survival in postmenopausal women. Clin Cancer Res. 2011;17(7):1867–1874. doi:10.1158/1078-0432.CCR-10-2021
3. Elebro K, Borgquist S, Simonsson M, et al. Combined androgen and estrogen receptor status in breast cancer: treatment prediction and prognosis in a population-based prospective cohort. Clin Cancer Res. 2015;21(16):3640–3650. doi:10.1158/1078-0432.CCR-14-2564
4. Peters AA, Buchanan G, Ricciardelli C, et al. Androgen receptor inhibits estrogen receptor-alpha activity and is prognostic in breast cancer. Cancer Res. 2009;69(15):6131–6140. doi:10.1158/0008-5472.CAN-09-0452
5. Park S, Koo JS, Kim MS, et al. Androgen receptor expression is significantly associated with better outcomes in estrogen receptor-positive breast cancers. Ann Oncol. 2011;22(8):1755–1762. doi:10.1093/annonc/mdq678
6. Robinson JL, Macarthur S, Ross-Innes CS, et al. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. EMBO J. 2011;30(15):3019–3027. doi:10.1038/emboj.2011.216
7. Van Itallie CM, Anderson JM. The molecular physiology of tight junction pores. Physiology. 2004;19:331–338. doi:10.1152/physiol.00027.2004
8. Singh AB, Dhawan P. Claudins and cancer: fall of the soldiers entrusted to protect the gate and keep the barrier intact. Semin Cell Dev Biol. 2015;42:58–65. doi:10.1016/j.semcdb.2015.05.001
9. Singh AB, Uppada SB, Dhawan P. Claudin proteins, outside-in signaling, and carcinogenesis. Pflugers Arch. 2017;469(1):69–75. doi:10.1007/s00424-016-1919-1
10. Gerhard R, Ricardo S, Albergaria A, et al. Immunohistochemical features of claudin-low intrinsic subtype in metaplastic breast carcinomas. Breast. 2012;21(3):354–360. doi:10.1016/j.breast.2012.03.001
11. Taylor NA, Vick SC, Iglesia MD, et al. Treg depletion potentiates checkpoint inhibition in claudin-low breast cancer. J Clin Invest. 2017;127(9):3472–3483. doi:10.1172/JCI90499
12. Meng J, Mostaghel EA, Vakar-Lopez F, Montgomery B, True L, Nelson PS. Testosterone regulates tight junction proteins and influences prostatic autoimmune responses. Horm Cancer. 2011;2(3):145–156. doi:10.1007/s12672-010-0063-1
13. Sutinen P, Malinen M, Heikkinen S, Palvimo JJ. SUMOylation modulates the transcriptional activity of androgen receptor in a target gene and pathway selective manner. Nucleic Acids Res. 2014;42(13):8310–8319. doi:10.1093/nar/gku543
14. Ashikari D, Takayama KI, Obinata D, Takahashi S, Inoue S. CLDN8, an androgen-regulated gene, promotes prostate cancer cell proliferation and migration. Cancer Sci. 2017;108(7):1386–1393. doi:10.1111/cas.13269
15. Offner S, Hekele A, Teichmann U, et al. Epithelial tight junction proteins as potential antibody targets for pancarcinoma therapy. Cancer Immunol Immunother. 2005;54(5):431–445. doi:10.1007/s00262-004-0613-x
16. Lu S, Singh K, Mangray S, et al. Claudin expression in high-grade invasive ductal carcinoma of the breast: correlation with the molecular subtype. Mod Pathol. 2013;26(4):485–495. doi:10.1038/modpathol.2012.187
17. Ni M, Chen Y, Fei T, et al. Amplitude modulation of androgen signaling by c-MYC. Genes Dev. 2013;27(7):734–748. doi:10.1101/gad.209569.112
18. Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi:10.1186/bcr2635
19. Feng J, Li L, Zhang N, et al. Androgen and AR contribute to breast cancer development and metastasis: an insight of mechanisms. Oncogene. 2017;36(20):2775–2790. doi:10.1038/onc.2016.432
20. Kensler KH, Poole EM, Heng YJ, et al. Androgen receptor expression and breast cancer survival: results from the nurses’ health studies. J Natl Cancer Inst. 2018;111:700–708.
21. Park S, Koo J, Park HS, et al. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21(3):488–492. doi:10.1093/annonc/mdp510
22. Castellano I, Allia E, Accortanzo V, et al. Androgen receptor expression is a significant prognostic factor in estrogen receptor positive breast cancers. Breast Cancer Res Treat. 2010;124(3):607–617. doi:10.1007/s10549-010-0761-y
23. Castellano I, Chiusa L, Vandone AM, et al. A simple and reproducible prognostic index in luminal ER-positive breast cancers. Ann Oncol. 2013;24(9):2292–2297. doi:10.1093/annonc/mdt183
24. Salvi S, Bonafe M, Bravaccini S. Androgen receptor in breast cancer: a wolf in sheep’s clothing? A lesson from prostate cancer. Seminars in Cancer Biology. 2020;60:132-137. doi: 10.1016/j.semcancer.2019.04.002.
25. Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi:10.1172/JCI45014
26. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
27. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
28. Dhawan P, Singh AB, Deane NG, et al. Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J Clin Invest. 2005;115(7):1765–1776. doi:10.1172/JCI24543
29. Osanai M, Takasawa A, Murata M, Sawada N. Claudins in cancer: bench to bedside. Pflugers Arch. 2017;469(1):55–67. doi:10.1007/s00424-016-1877-7
30. Pope JL, Bhat AA, Sharma A, et al. Claudin-1 regulates intestinal epithelial homeostasis through the modulation of Notch-signalling. Gut. 2014;63(4):622–634. doi:10.1136/gutjnl-2012-304241
31. O’Leary KA, Rugowski DE, Sullivan R, Schuler LA. Prolactin cooperates with loss of p53 to promote claudin-low mammary carcinomas. Oncogene. 2014;33(23):3075–3082. doi:10.1038/onc.2013.278
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.