Back to Journals » Infection and Drug Resistance » Volume 12
The evaluation of the synergistic antimicrobial and antibiofilm activity of AamAP1-Lysine with conventional antibiotics against representative resistant strains of both Gram-positive and Gram-negative bacteria
Authors Almaaytah A, Abualhaijaa A, Alqudah O
Received 9 February 2019
Accepted for publication 10 April 2019
Published 23 May 2019 Volume 2019:12 Pages 1371—1380
DOI https://doi.org/10.2147/IDR.S204626
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Ammar Almaaytah,1 Ahmad Abualhaijaa,2 Obadah Alqudah3
1Department of Pharmaceutical Technology, Faculty of Pharmacy, Jordan University of Science and Technology, Irbid, Jordan; 2Department of Applied Biological Sciences, Faculty of Science and Arts, Jordan University of Science and Technology, Irbid, Jordan; 3Department of Legal Medicine, Toxicology and Forensic Medicine, Faculty of Medicine, Jordan University of Science and Technology, Irbid, Jordan
Background and purpose: Antimicrobial resistance toward antibiotics is reaching historical unprecedented levels. There is an urgent and imminent need to develop novel antimicrobial alternatives. Antimicrobial peptides could prove to be a successful group of antimicrobials for drug development. Recently, we have designed a novel synthetic peptide named AamAP1-Lysine. The peptide displayed potent wide-spectrum antimicrobial activities against Gram-positive and Gram-negative bacteria. The purpose of this study is to evaluate the antimicrobial effect of combining AamAP1-Lysine with five different conventional antibiotics each representing a distinct mechanism of action in order to explore the possibility of producing a synergistic mode of action against a resistant strain of Gram-positive and a resistant strain of Gram-negative bacteria.
Methodology: The antimicrobial activity of AamAP1-Lysine in combination with five different antibiotics were evaluated for their antimicrobial activity employing standard antimicrobial assays, the synergistic activity of the peptide-antibiotic combinations were evaluated using checkerboard technique in addition to real-time time-kill assays. For the antibiofilm studies, the MBEC values were determined by employing the Calgary device.
Results: The combination strategy displayed potent synergistic activities against planktonic bacteria in a significant number of peptide-antibiotic combinations. The synergistic activity managed to reduce the effective minimum inhibitory concentration (MIC) concentrations dramatically with some combinations exhibiting a 64-fold decrease in the effective MIC of AamAP1-Lysine individually. Additionally, the combined synergistic activities of the peptide antibiotics were evaluated, and our results have identified two peptide antibiotic combinations with potent synergistic activities against biofilm growing strains of resistant bacteria.
Conclusion: Our results clearly indicate that peptide-antibiotic combinations could prove to be a very effective strategy in combatting multidrug-resistant bacteria and biofilm caused infections.
Keywords: antimicrobial peptides, AamAP1-Lysine, bacterial resistance, antibiofilm, synergy
Introduction
Recent global health reports have displayed a significant rise in the number of infections that are caused by resistant microorganisms against a wide range of antibiotics.1 These reports indicate that the current medical community is facing a critical time in human history where most clinically developed antibiotics will be rendered obsolete in the face of microbial infections.2 This situation is challenging humanity with the possibility of humans entering the post-antibiotic era and thus rendering simple surgical procedures and infections that once were treated with the routine administration of antibiotics to become severely life threatening.3 Current biomedical scientists and medical experts are continuously seeking the development of novel classes of antimicrobial agents to tackle the issue of microbial resistance and to rejuvenate the dry pipelines of antimicrobial drug development and discovery faced currently by the pharma industry.4 One of the highly researched and potential classes of antimicrobial agents that have been studied in the previous decade for successful antimicrobial development are antimicrobial peptides (AMPs).5 This group of molecules are usually composed on average of 5–50 amino acids and carry a net positive charge while exhibiting an amphipathic nature, structural features that collectively play an important role in disrupting bacterial membranes and eventually causing cell lysis and death.6 Although several hundred AMPs have been characterized from natural sources or through de-novo synthetic drug design and were reported to exhibit potent antimicrobial activities, most of these molecules faced several obstacles that hampered their clinical development due to their toxicity against mammalian cells.7 Several studies have tried to bypass this issue through different approaches including sequence modification, peptide hybridization and the use of D-natural amino acid substitution to maximize the selectivity of the peptides toward bacterial cells and consequently increase their selectivity index.8,9 Recently, we have designed a novel synthetic peptide analog named AamAP1-Lysine.10 The peptide was designed based on lysine amino acid substitution of a natural scorpion derived AMP in order to enhance its activity and reduce its overall cytotoxicity. The design strategy managed to produce a peptide analog with a tenfold increase in potency and a significant enhancement of the overall selectivity index when compared with the parent peptide. The aim of this study is to explore the benefit of combining AamAP1-Lysine with conventional antibiotics to produce a synergistic mode of action against a representative sample of resistant strains of Gram-positive and Gram-negative bacteria. This approach could further increase the selectivity index of the peptide and enhance its overall toxicity profile for the purpose of antimicrobial drug development. Additionally, this combination strategy would also be evaluated for inhibiting biofilm formation for the same strains involved in the synergistic studies.
Materials and methods
Materials
AamAP1-Lysine is an 18-residue amino acid (FLFKLIPKAIKKLISKFK) synthetic peptide analog that was designed previously in-house using a naturally identified scorpion AMP as a template for peptide rational design. The peptide displays an overall charge of +6 and displays a hydrophobic moment and a hydrophobicity average of (0.61), respectively. The peptide was synthesized using solid-phase Fmoc chemistry and purified to (>95%) purity with its identity confirmed using ESI-MS spectrometry (G.L Biochem, People’s Republic of China). The five antibiotics employed for the synergy studies including levofloxacin, ampicillin, chloramphenicol, rifampicin and erythromycin were purchased commercially from Sigma Aldrich. The antibiotics and their stock solutions were reconstituted according to the manufacturer’s instructions and frozen at −80°C.
Microorganisms
The bacterial strains employed in both the antimicrobial and antibiofilm synergistic studies were acquired from the American Tissue Culture Collection (ATCC), with representatives of both control and resistant Gram-positive and Gram-negative bacteria. The strains included two Gram-positive strains of Staphylococcus aureus (ATCC 29213) as a control strain and the methicillin-resistant Staphylococcus aureus (ATCC 33591). For Gram-negative bacteria, two bacterial strains were employed in the study and these include Pseudomonas aeruginosa (ATCC 27853) and the multidrug-resistant strain of Pseudomonas aeruginosa (ATCC BAA 2114).
Antimicrobial susceptibility assay
The determination of the antimicrobial activity of both AamAP1-Lysine and the antibiotics included in this study was performed according to the microbroth dilution method as outlined by the Clinical and Laboratory Standards Institute.11,12 Briefly, all bacterial strains were grown overnight in Muller Hinton Broth (MHB) and diluted the following day to reach a concentration of 106 CFU/mL. This was followed by serial dilution of AamAP1-Lysine and the antibiotics included in the study for the purpose of preparing different concentrations of each antimicrobial component. The peptide and the antibiotics were later loaded on a 96-well mL plates at a volume of 50 μL and mixed appropriately with the relevant microorganism to reach a final bacterial concentration of 5×105 CFU/mL. The plates were then incubated for 24 at hours in a humidified incubator at 37°C and the bacterial growth inhibitory activity represented by the minimum inhibitory concentration (MIC) was determined through optical measurement using an ELISA plate reader (Biotek, Canada).
Synergistic studies and checkerboard assay
The synergetic activity of AamAP1-Lysine- antibiotic combinations were evaluated by the determination of the fractional inhibitory concentration (FIC) and as performed previously.13,14 The broth microdilution checkerboard technique was employed for FIC determination, with the FIC index defined as the sum of each MIC of the peptide-antibiotic combination divided by each individual antimicrobial agent and according to the following equation:
FIC index = (MIC of drug X in combination) / (MIC of drug X alone) + (MIC of drug Y in combination) / (MIC of drug Y alone). FIC indices were interpreted as follows: £0.5: synergistic activity, 0.5–1: additive activity, 1–4: indifference, >4: antagonism
Time-kill assays
In order to confirm the synergistic mode of killing for the peptide-antibiotic combinations that were obtained from the checkerboard assay, time-dependent kill assays were carried out as performed previously.15–18 Bacterial strains were grown overnight in MHB at 37°C and later diluted to 1×106 CFU/mL with fresh MHB. The diluted cultures were incubated with the synergistic peptide-antibiotic combinations for 24 hrs at 37°C and aliquots of 0.1 mL were removed at (0, 2, 4, 8 and 24 hrs) time intervals, serially diluted and plated for viable count determination. Synergy was defined as a ≥2 log10 decrease in the CFU/mL below the initial starting inoculum or the most active individual component.
The time-kill assays for all experiments were performed at least thrice for confirmation of the results; the data are represented as mean ± standard deviation.
Bacterial biofilm formation
Bacterial biofilm formation was performed as described previously.19–21 Briefly, the control and resistant bacterial strains of both S. aureus and P. aeruginosa were grown in MHB for 20 hrs at 37°C followed by dilution in the same medium to achieve a total concentration of 107 CFU/mL. The biofilm 96 peg-lids were employed for biofilm growth and accumulation and 150 μL of each bacterial culture was loaded on these peg-lids accordingly. The Calgary device was incubated for 20 hrs under continuous rotation at 125 rpm and 37°C and as described by the manufacturer to allow for biofilm formation.
Synergistic antibiofilm assay
The Calgary device described previously was employed for the determination of the minimum biofilm eradication concentration (MBEC) which is defined as the minimum concentration of an antimicrobial agent needed to inhibit the re-growth of biofilms after exposure to an antimicrobial agent. Briefly and as described in previously.22,23 PBS was employed to wash all planktonic cells on the device related biofilm pegs. This was followed by transferring the pegs into a 96-well challenge plate containing all the antimicrobial agents and the combinations that were employed in the assay. MBEC was determined using an ELISA reader at λ=550 nm. FIC was determined as described in the previous assay.
Results
Synthesis of AamAP1-lysine
AamAP1-Lysine was synthesized according to conventional solid-phase Fmoc chemistry. The peptide purity was evaluated to be over 95% as indicated by HPLC analysis (Figure S1). The identity of the peptide was confirmed by ESI-MS with the synthetic peptide displaying major peaks in the +2, +3, +4 and +5 charge state of 1053.70, 702.85 , 527.40 and 422 Da (Figure S2).
Antimicrobial and synergistic checkerboard assays
Table 1 lists the MIC results of all the antimicrobial agents against the representative strains of bacterial strains that were employed in the study. The combination of AamAP1-Lysine and the antibiotics resulted in significant enhancement of the antimicrobial activity of many peptide-antibiotic combinations (Table 2). The MIC values for these groups decreased dramatically as six peptide-antibiotic combinations displayed significant synergistic activities. The combinations that proved to exhibit synergistic activities against the control strain S. aureus (ATCC 29213) included AamAP1-Lysine-rifampicin and the AamAP1-Lysine-erythromycin combination. For the resistant strain of S. aureus (ATCC 33591), AamAP1-Lysine-levofloxacin and AamAP1-Lysine-erythromycin displayed potent synergistic effects. For Gram-negative bacteria, no synergistic activities were reported for the control strain of P. aeruginosa (ATCC 27853). However, both AamAP1-Lysine-levofloxacin and AamAP1-Lysine-chloramphenicol managed to synergistically inhibit the growth of the MDR strain of P. aeruginosa (BAA2114) with FIC indices of 0.36 for both combinations (Table 3). The most potent synergistic combinations as represented by their FIC index belonged to AamAP1-Lysine-levofloxacin and AamAP1-Lysine-rifampicin against the resistant strain of S. aureus (ATCC 33591) with FIC values of 0.103 and 0.128, respectively. The rest of the peptide antibiotic combinations displayed additive effects with FIC indices in the range of (0.5<FIC<1). The only peptide-antibiotic combination exhibiting antagonistic activity was AamAP1-Lysine-erythromycin against the resistant strain of S. aureus (ATCC 33591) Table 3 Lists all the synergistic combinations employed in the studies and their FIC indices accordingly.
 | Table 1 Minimum inhibitory concentrations (MICs) of AamAP1-Lysine and the antibiotics individually against the tested bacterial strains. Results represent triplicates |
 | Table 2 Minimum inhibitory concentrations (MICs) of AamAP1-Lysine and the antibiotics in combination against Gram-positive and Gram-negative bacteria. Results represent triplicates |
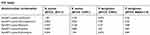 | Table 3 The fractional inhibitory concentration (FIC) indices of the antimicrobial combinations. |
Synergistic time-kill assays
Time-kill assays were performed in order to confirm the synergistic results that were obtained from the checkerboard assays. In all synergistic combinations at ½ of their effective MICs, none of the individual antimicrobial agents including AamAP1-Lysine and the antibiotics managed to induce cell death within 24-hr exposure to the individual antimicrobial agents (Figure 1). However, when combined together, all of the synergistic combinations including AamAP1-Lysine-ampicillin and AamAP1-Lysine-erythromycin managed within 4 hrs of exposure to significantly reduce the bacterial counts of the control strain of S. aureus (ATCC 29213). Complete elimination of bacteria was achieved after an 8 hrs interval time. For the resistant strain of S. aureus ATCC (33591), both AamAP1-Lysine-levofloxacin and AamAP1-Lysine-rifampicin managed to follow the same pattern as with the control strain and in confirmation with the synergistic checkerboard results. For Gram-negative bacteria represented by the resistant strain of P. aeruginosa ATCC (BAA2114), the AamAP1-Lysine-levofloxacin and the AamAP1-Lysine-chloramphenicol combination caused a marked reduction in viable colony count within 2 hrs of peptide antibiotic exposure and complete eradication of bacteria was reported at 8 hrs of exposure. Bactericidal activity (≥3 log10 CFU/mL) was achieved within 2–4 hrs against all bacterial strains (Figure 1). These results in addition to the checkerboard assay clearly display the synergistic potential of the successful peptide-antibiotic combinations in killing the representative bacterial strains that were employed in the study.
Antibiofilm combination studies
MBEC was employed to assess the antibiofilm activity of the individual antimicrobial agents including AamAP1-Lysine and the antibiotics in addition to evaluating their combinatorial synergistic effects. MBEC values for both S. aureus (ATCC 33591) and P. aeruginosa (ATCC BAA2114) were 60 and 70 μM, respectively. Of all the peptide-antibiotic combinations against both resistant strains of Gram-positive and Gram-negative bacteria, only two combinations managed successfully to produce a synergistic antibiofilm mode of action. AamAP1-Lysine in combination with rifampicin exhibited potent synergistic activity against S. aureus (ATCC 33591) with a FIC index of 0.058 representing a dramatic 60-fold decrease in the MBEC value of AamAP1-Lysine individually (Table 4). For the MDR P. aeruginosa (ATCC BAA2114) the synergistic mode of action was reported for the levofloxacin peptide combination which resulted in a 28-fold reduction in AamAP1-Lysine’s MBEC value (Table 4).
 | Table 4 The MBEC values and the FIC index of the successful synergistic combinations of AamAP1-Lysine combined with antibiotics |
Discussion
The escalating global dilemma of microbial resistance against antibiotics is putting a huge stress on global health authorities to accelerate the development of antibiotic alternatives in order to tackle this serious health issue and contribute to saving human lives.24 Bacterial resistance is growing in numbers with some microbial strains displaying pan-resistance against all conventional antibiotics employed in the clinic and threatening the overall health of mankind.25 Additionally, bacteria and their ability to form biofilms with its characteristic inherent resistance are posing an additional threat to human health as several infections including wound and catheter-related infections are directly linked to biofilms spread and their related morbidities.26 These critical health threatening situations require urgent innovative and successful therapeutic approaches.
During the last two decades, several AMPs have been characterized and developed as an alternative approach to conventional antibiotics.27 Despite having significant efficacy in eradicating a wide spectrum of resistant microorganisms, AMPs faced several obstacles in being developed into successful therapeutics. One of the major drawbacks related to this group of molecules is related to their lack of specific target selectivity.28 This lack of apparent selectivity increases the toxicity of these peptides against normal non-bacterial cells and consequently is hampering the development of these agents as alternatives to conventional antibiotics. Recently, we have designed a synthetic novel AMP named AamAP1-Lysine with potent broad-spectrum antimicrobial activity against different strains of Gram-positive and Gram-negative bacteria. Several methods have been employed to minimize and control the toxicity of AMPs including peptide sequence modification, nano-encapsulation and peptide hybridization.8,9,28 The aim of this study was to assess the potential of combining AamAP1-Lysine with conventional antibiotics in order to achieve a synergistic mode of antibacterial activity that would decrease the individual MIC of each individual antibiotic and eventually increase their therapeutic index. According to our study, AamAP1-Lysine displayed significant synergistic antimicrobial and antibiofilm effects against a sample of representative strains of control and resistant Gram-positive and Gram-negative bacteria when combined with conventional antibiotics. The checkerboard assays revealed that the peptide is displaying synergistic activity against planktonic cells of both control and resistant strains of S. aureus (ATCC 29213, ATCC 33591) and P. aeruginosa (ATCC 27853, ATCC BAA 2114). For the planktonic Gram-positive bacteria, three peptide-antibiotic combinations proved to be highly synergistic and these include levofloxacin, rifampicin and erythromycin. The levofloxacin combination proved to be the most potent with a FIC index of 0.103 against the resistant strain of S. aureus (ATCC 33591) and in accordance synergistic studies performed previously.29–31 For Gram-negative bacteria, only two combinations displayed a synergistic activity, and these include levofloxacin and chloramphenicol. The two combinations had equal synergistic activity with a FIC index of 0.36 against the resistant form of P. aeruginosa (ATCC BAA 2114). Additionally, the combination strategy proved to be effective in inhibiting biofilm formation as indicated by the dramatic reduction in the MBEC values for the successful synergistic combinations. When combined with chloramphenicol, AamAP1-Lysine managed to inhibit biofilm growth for S. aureus (ATCC 33591) at 1 μM displaying a 60-fold decrease in the MBEC value when compared with the individual peptide. For the MDR form of P. aeruginosa, AamAP1-Lysine combined with levofloxacin managed to decrease the MBEC value of the peptide by 28-fold with a FIC index of 0.11. These results clearly display that combining antibiotics with AamAP1-Lysine is an effective strategy in enhancing the antimicrobial activity of AMPs against both planktonic and biofilm forms of bacteria.
The dramatic decrease in the MIC values of AamAP1-Lysine as a result of the synergistic action of the combination strategy could prove to be a very useful strategy for AMPs clinical development. The reduction in the MIC value will diminish the toxicity of this group of molecules against normal mammalian cells and allow these peptides to be employed at significantly at reduced concentrations and thus solving the issue of AMPs toxicity in addition to cutting the cost of manufacturing these peptides on a large scale due to their enhanced potency. This strategy could also be employed to AMPs that are currently being used in the clinic such as colistin, this AMP is reserved only for unresponsive and highly resistant Gram-negative bacterial infections as a result of its renal toxicity and significant potency. However, recent reports have identified several strains in China, USA and Europe with acquired resistance against colistin rendering this last resort drug highly fragile against bacterial resistance.32–34 Combining antibiotics with colistin to achieve synergistic outcomes could allow clinicians to avoid this challenge and extend the lifespan of such antibiotics. The mode of action of the synergistic activity is still unclear and further molecular mechanistic studies are required to explain the underlying mechanism of action responsible for such enhanced antimicrobial activity. One theory suggests that AMPs with their capability of creating transient pores within membranes of bacterial cells could facilitate the entry of conventional antibiotics intracellularly and achieve enhanced antimicrobial outcomes.
In conclusion, we report the evaluation and characterization of the antimicrobial and antibiofilm activity of AamAP1-Lysine in combination with conventional antibiotics against representative resistant strains of Gram-positive and Gram-negative bacteria. Our results proved that the combination strategy resulted in dramatic decrease of the effective concentrations of AamAP1-Lysine needed to inhibit several representative strains of both planktonic and biofilm forms of bacteria and consequently reduce its toxicity. These results proved that AamAP1-Lysine in combination with conventional antibiotics could prove to be a very attractive candidate for antimicrobial drug development.
Acknowledgments
This work was carried out with the support of the Deanship of Research at Jordan University of Science and Technology (project No. 136/2015).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ferri M, Ranucci E, Romagnoli P, Giaccone V. Antimicrobial resistance: a global emerging threat to public health systems. Crit Rev Food Sci Nutr. 2017;57(13):2857–2876. doi:10.1080/10408398.2015.1077192
2. Munita JM, Shelburne S, Greenberg DE, Arias CA. The GROWING THREAT OF ANTIMICROBIAL RESISTANCE. Tex Med. 2017;113(2):48–52.
3. Jones CA, Davis JS, Looke DF. Death from an untreatable infection may signal the start of the post-antibiotic era. Med J Aust. 2017;206(7):292–293.
4. Luepke KH, Suda KJ, Boucher H, et al. Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy. 2017;37(1):71–84. doi:10.1002/phar.1868
5. Mishra B, Reiling S, Zarena D, Wang G. Host defense antimicrobial peptides as antibiotics: design and application strategies. Curr Opin Chem Biol. 2017;38:87–96. doi:10.1016/j.cbpa.2017.03.014
6. Bechinger B, Gorr S-U. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res. 2017;96(3):254–260. doi:10.1177/0022034516679973
7. Mahlapuu M, Håkansson J, Ringstad L, Camilla B. Antimicrobial peptides: an emerging category of therapeutic agents. Front Cell Infect Microbiol. 2016;6:194. doi:10.3389/fcimb.2016.00194
8. Cardoso MH, Cândido ES, Oshiro KGN, Rezende SB, Franco OL. Peptides containing d-amino acids and retro-inverso peptides: general applications and special focus on antimicrobial peptides. In: Peptide Applications in Biomedicine, Biotechnology and Bioengineering. Woodhead Publishing. 2018:131–155.
9. Almaaytah A, Tarazi S, Al-Fandi M, Abuilhaija A, Al-Balas Q, Abu-Awad A. The design and functional characterization of the antimicrobial and antibiofilm activities of BMAP27-melittin, a rationally designed hybrid peptide. Int J Pept Res Ther. 2015;21(2):165–177. doi:10.1007/s10989-014-9444-6
10. Almaaytah A, Tarazi S, Abu-Alhaijaa A, et al. Enhanced antimicrobial activity of AamAP1-Lysine, a novel synthetic peptide analog derived from the scorpion venom peptide AamAP1. Pharmaceuticals. 2014;7(5):502–516. doi:10.3390/ph7050502
11. Ouhara K, Komatsuzawa H, Kawai T, et al. Increased resistance to cationic antimicrobial peptide LL-37 in methicillin-resistant strains of Staphylococcus aureus. J Antimicrob Chemother. 2008;61(6):1266–1269. doi:10.1093/jac/dkn106
12. Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc. 2008;3(2):163. doi:10.1038/nprot.2007.521
13. Sueke H, Kaye SB, Neal T, Hall A, Tuft S, Parry CM. An in vitro investigation of synergy or antagonism between antimicrobial combinations against isolates from bacterial keratitis. Invest Ophthalmol Vis Sci. 2010;51(8):4151–4155. doi:10.1167/iovs.09-4839
14. Beckloff N, Laube D, Castro T, et al. Activity of an antimicrobial peptide mimetic against planktonic and biofilm cultures of oral pathogens. Antimicrob Agents Chemother. 2007;51(11):4125–4132. doi:10.1128/AAC.00208-07
15. Zheng -Y-Y, Du R-L, Cai S-Y, et al. Study of benzofuroquinolinium derivatives as a new class of potent antibacterial agent and the mode of inhibition targeting FtsZ. Front Microbiol. 2018;9:1937. doi:10.3389/fmicb.2018.01937
16. Yoon J, Urban C, Terzian C, Mariano N, Rahal JJ. In vitro double and triple synergistic activities of polymyxin B, imipenem, and rifampin against multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother. 2004;48(3):753–757.
17. Mun S-H, Joung D-K, Kim Y-S, et al. Synergistic antibacterial effect of curcumin against methicillin-resistant Staphylococcus aureus. Phytomedicine. 2013;20(8–9):714–718. doi:10.1016/j.phymed.2013.02.006
18. Luca V, Stringaro A, Colone M, Pini A, Mangoni ML. Esculentin (1-21), an amphibian skin membrane-active peptide with potent activity on both planktonic and biofilm cells of the bacterial pathogen Pseudomonas aeruginosa. Cell Mol Life Sci. 2013;70(15):2773–2786. doi:10.1007/s00018-013-1291-7
19. Almaaytah A, Farajallah A, Abualhaijaa A, Al-Balas Q. A3, a scorpion venom derived peptide analogue with potent antimicrobial and potential antibiofilm activity against clinical isolates of multi-drug resistant gram positive bacteria. Molecules. 2018;23(7):1603. doi:10.3390/molecules23071603
20. Feng X, Sambanthamoorthy K, Palys T, Paranavitana C. The human antimicrobial peptide LL-37 and its fragments possess both antimicrobial and antibiofilm activities against multidrug-resistant acinetobacter baumannii. Peptides. 2013;49:131–137. doi:10.1016/j.peptides.2013.09.007
21. Schwering M, Song J, Louie M, Turner RJ, Ceri H. Multi-species biofilms defined from drinking water microorganisms provide increased protection against chlorine disinfection. Biofouling. 2013;29(8):917–928. doi:10.1080/08927014.2013.816298
22. Almaaytah A, Qaoud MT, Mohammed GK, et al. Antimicrobial and antibiofilm activity of UP-5, an ultrashort antimicrobial peptide designed using only arginine and biphenylalanine. Pharmaceuticals. 2018;11(1):3. doi:10.3390/ph11010003
23. Littmann J, Buyx A, Cars O. Antibiotic resistance: an ethical challenge. Int J Antimicrob Agents. 2015;46(4):359–361. doi:10.1016/j.ijantimicag.2015.06.010
24. Paterson DL, Harris PNA. Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis. 2016;16(2):132–133. doi:10.1016/S1473-3099(15)00463-6
25. Das T, Kimyon O, Manefield MJ. Bacterial biofilms on wounds, a major factor that delays wound healing and a potential threat to human life and economy In: Recent Clinical Techniques, Results, and Research in Wounds. Springer, Cham. 2017;1–20. doi:10.1007/15695_2017_6.
26. Wang G, editor. Antimicrobial Peptides: Discovery, Design and Novel Therapeutic Strategies. Cabi; Oxfordshire, UK.2017.
27. Narayana JL, Chen J-Y. Antimicrobial peptides: possible anti-infective agents. Peptides. 2015;72:88–94. doi:10.1016/j.peptides.2015.05.012
28. Wang H, Kaijin X, Liu L, et al. The efficacy of self-assembled cationic antimicrobial peptide nanoparticles against cryptococcus neoformans for the treatment of meningitis. Biomaterials. 2010;31(10):2874–2881. doi:10.1016/j.biomaterials.2009.12.042
29. Almaaytah A, Alnaamneh A, Abualhaijaa A, Al-Balas Q. In vitro synergistic activities of the hybrid antimicrobial peptide MelitAP-27 in combination with conventional antibiotics against planktonic and biofilm forming bacteria. Int J Pept Res Ther. 2016;22(4):497–504. doi:10.1007/s10989-016-9530-z
30. Fang Z, Ban L, Yanan L, et al. A quinoline-based FtsZ inhibitor for the study of antimicrobial activity and synergistic effects with β-lactam antibiotics. J Pharmacol Sci. 2018;137(3):283–289. doi:10.1016/j.jphs.2018.07.005
31. Tan CM, Therien AG, Lu J, et al. Restoring methicillin-resistant Staphylococcus aureus susceptibility to β-lactam antibiotics. Sci Transl Med. 2012;4(126):126ra35–126ra35. doi:10.1126/scitranslmed.3003592
32. Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
33. Mediavilla JR, Patrawalla A, Chen L, et al. Colistin-and carbapenem-resistant Escherichia coli harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States. MBio. 2016;7(4):e01191–16. doi:10.1128/mBio.01191-16
34. Skov RL, Monnet DL. Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Eurosurveillance. 2016;21(9):30155. doi:10.2807/1560-7917.ES.2016.21.9.30155
Supplementary material
 | Figure S1 High-performance liquid chromatography analysis of AamAP1-Lysine. |
 | Figure S2 ESI-MS analysis of AamAP1-Lysine identifying displaying major peaks in the +2, +3, +4 and +5 charge state of 1053.70 , 702.85 , 527.40 and 422 Da, respectively. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

