Back to Journals » Cancer Management and Research » Volume 11
The emerging roles of PIWI-interacting RNA in human cancers
Received 21 March 2019
Accepted for publication 14 May 2019
Published 28 June 2019 Volume 2019:11 Pages 5895—5909
DOI https://doi.org/10.2147/CMAR.S209300
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Yaya Yu,1 Jing Xiao,2 Swei Sunny Hann1,3
1Laboratory of Tumor Biology, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong Province, 510120, People’s Republic of China; 2Department of Gynecology, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong Province, 510120, People’s Republic of China; 3Guangdong Provincial Key Laboratory of Clinical Research on Traditional Chinese Medicine Syndrome, The Second Clinical College of Guangzhou University of Chinese Medicine, Guangzhou, Guangdong Province, 510120, People’s Republic of China
Abstract: PIWI-interacting RNAs (piRNAs) are a type of non-coding RNAs that interact with PIWI proteins, which are members of the argonaute family. Originally described in the germline, piRNAs are also expressed in human somatic cells in a tissue-specific manner. piRNAs are involved in spermatogenesis, germ stem-cell maintenance, silencing of transposon, epigenetic and genomic regulation and rearrangement. A large number of studies have demonstrated that expression of piRNAs is involved in many kinds of disease, including cancer. Abnormal expression of piRNAs is emerging as a critical player in cancer cell proliferation, apoptosis, invasion, and migration in vitro and in vivo. Functionally, piRNAs maintain genomic integrity by repressing the mobilization of transposable elements, and regulate the expression of downstream target genes via transcriptional or post-transcriptional mechanisms. Furthermore, altered expression of piRNAs in cancer is linked to clinical outcome, highlighting the important role that they may play as novel diagnostic and prognostic biomarkers, and as therapeutic targets for cancer therapy. In this review, we focus on the biogenesis and the functional roles of piRNAs in cancers, discuss emerging insights into the roles of piRNAs in the occurrence, progression, and treatment of cancers, reveal various mechanisms underlying piRNAs-mediated gene regulation, and highlight their potential clinical utilities as biomarkers as well as potential targets for cancer treatment.
Keywords: PIWI-interacting RNA, cancer, biogenesis, biomarkers, therapeutics
Introduction
Among human genes, 70% comprise the actively transcribed genome, less than 3% are protein-coding genes, and most of them are non-coding RNAs (ncRNAs).1–3 The discovery of ncRNAs provides us with a brand new way of understanding gene expression and regulation. Unlike messenger RNA (mRNA), ncRNA functions either in the nucleus by binding to DNA to contribute to gene silencing, or in the cytoplasm by regulating mRNA to affect protein expression. The machinery of coding RNAs and ncRNAs works together to guarantee overall homeostasis. There are two kinds of ncRNA, with different functions, namely regulatory ncRNAs and housekeeper ncRNAs. Regulatory ncRNAs can be further divided into long non-coding RNAs (lncRNAs) and small non-coding RNAs (sncRNAs) according to transcript size. PIWI-interacting RNAs (piRNAs), microRNAs (miRNAs), small interfering RNAs (siRNAs), and small nucleolar RNAs (snoRNAs) are well-known types of sncRNA.4–10
PIWI-interacting RNAs (piRNAs), a class of sncRNA molecules with the size of 24–32 nucleotides, are associated with the PIWI proteins, which are a subfamily of argonaute proteins.11,12 More than 30,000 piRNAs have been discovered within the eukaryotic genome, including the human genome.13,14 piRNAs were considered to be the “dark matter” of ncRNAs in the past decade. However, more recent evidence has shown their biological and clinical potential as emerging studies demonstrate that aberrant expression of piRNAs is a unique characteristic in several diseases, including cancers.15–17 It has been found that piRNAs can distinguish tumor from non-tumor, and are involved in cancer cell proliferation, apoptosis, invasion, and migration both in vitro and in vivo.17–22 Furthermore, the altered expression of piRNAs is strongly associated with clinical outcome, highlighting their important role in cancer. piRNAs may serve as novel diagnostic and prognostic biomarkers, and as effective targets for cancer therapy.17–22 piRNAs could repress the mobilization of transposable elements (TEs) and regulate the expression of downstream target genes to maintain genomic integrity via transcriptional or post-transcriptional mechanisms. Moreover, an increasing amount of evidence has shown that piRNAs have either oncogenic or tumor-suppressive properties in cancer.17,23 However, the underlying tumorigenic functions and potential clinical value of piRNAs remain elusive and require further elucidation. In this review, we focus on the biogenesis of piRNAs and the emerging functional role of piRNAs in a variety of cancers, and discuss potential diagnostic and prognostic utilities of piRNAs in cancer.
Discovery of piRNAs
First discovered in 2006, four groups independently identified this important class of small RNAs from mouse male germ cells.24,25 With the help of techniques, such as immunoprecipitation and northern blot methods, piRNAs were also detected in human ovaries and testes.26 Several features of piRNAs in mice and humans have been reported. They: 1) are mainly present in germ cells; 2) are expressed in a regulated manner; 3) map to the genome via clusters; 4) show a strong tendency for uridine (U) at the 5′ end; 5) have both sense and antisense directions; 6) are missing stem loop structures; 7) have 5′-phosphate and 3′-OH groups; 8) Dicer biogenesis is independent of piRNAs; and 9) interact with PIWI proteins. They are one of the most abundantly expressed small RNAs, with every human spermatid containing approximately 1 million piRNAs. In the mouse, the PIWI proteins are expressed temporally during spermatogenesis, indicating a time-dependent expression for piRNAs.
There are three groups of piRNAs, namely lncRNA-derived, mRNA-derived, and transposon-derived piRNAs.27,28 The entire lncRNA transcript is the typical source of lncRNA-derived piRNAs. mRNA-derived piRNAs originate from the 3ʹ-untranslated regions (UTRs) of mRNAs and are sense to the mRNA from which they are processed. The transposon-derived piRNAs come from two genomic strands and produce both sense and antisense piRNAs.27,28 Among these three piRNAs, only the function of transposon-derived piRNAs has been relatively well studied. In humans, a few piRNAs are mapped to the TE, while most of them are mapped to the intergenic regions, indicating that regulation of transposons may not be the primary function of piRNAs. One study29 profiled the piRNAs from human adult testis and found that both lncRNAs and the genic regions of protein-coding genes could be sources of piRNAs in humans. The latter include the coding regions, 3′-UTRs and 5′-UTRs, while 3′-UTRs are the enriched origin of piRNAs. The levels of piRNAs were not strongly associated with the expression of the corresponding host genes, implying an additional mechanism for piRNA generation from protein-coding genes. Other studies found that transfer RNAs (tRNAs) and snoRNAs may be sources of piRNAs, as some piRNA genes existed within the boundaries of snoRNAs and tRNAs.30–32
Biogenesis of piRNAs
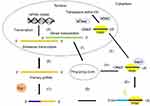
The biogenesis pathway of piRNAs is derived from the Drosophila pathway. The biogenesis of piRNAs is associated with silencing of target genes and requires three PIWI proteins, namely PIWI, Argonaute-3 (AGO3), and Aubergine (Aub). PIWI proteins are mainly located in the nuclei of somatic cells and germ cells,33 while AGO3 and AUB are observed in the cytoplasm of germ cells.34 AGO3 has an preference for an adenine (A) at the 10th sequence position, mapping sense to transposons, whereas AUB and PIWI prefer sequences with a 5′-U bias, mapping antisense to transposons.34 Unlike miRNAs, the biogenesis of piRNAs requires neither a double-stranded precursor nor the Dicer enzyme.35,36 In addition, piRNAs have not shown an overlap or any phasing within a cluster sequence.37 However, the precursors of piRNAs and other sncRNAs need more post-transcriptional processing to become mature sncRNAs. There are two distinct cytoplasmic synthesis pathways for the putative precursors to become fully mature, one of which is known as the “ping-pong” amplification pathway.27,28 The primary piRNAs are defined by the endonuclease Zucchini (Zuc), which was observed in germ and somatic cells, while biogenesis of secondary piRNAs was found in germ cells depending upon piRNA-guided reciprocal cleavage of sense and antisense transcripts, which resulted in piRNA amplification (ping-pong cycle).38 This is crucial for maintaining the levels of piRNAs and silencing of target genes.
In the nucleus, primary transcripts of piRNAs are first cleaved by Zuc, a riboendonuclease enzyme, generating a 5ʹ-phosphate residue. Then, the 3ʹ fragment of the transcripts is incorporated with PIWI and a 3ʹ to 5ʹ-exonuclease trims the transcripts to their final length. The 2ʹ-hydroxy group at the 3ʹ end is methylated by Hen1. The 5ʹ-end residue of the piRNAs incorporated with PIWI shows a strong bias for U residues. After being exported into the cytoplasmic production centers, the cluster transcripts are processed into smaller sequences and reach their partners to form piRNA–PIWI complexes.39 Then, the complexes migrate to the nucleus again and block the transcription of the target gene. Thus, piRNAs are transcriptional regulators that act mainly on TE sequences by recruiting histone methyltransferases.37,40 The process of piRNA biogenesis suggests that the size of mature piRNAs is a consequence of loading of piRNA intermediates into PIWI,34,41 followed by trimming through an unknown exonuclease or 3ʹ-end formation by Zuc.42,43 The primary mechanism of piRNA synthesis is shown in Figure 1.
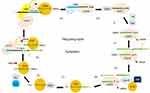
The ping-pong amplification loop of piRNAs
Discovered in Drosophila, the ping-pong cycle serves the purposes of generating piRNAs and silencing transposons in the cytoplasm.15 Differently from the primary synthesis of joining with PIWI to form piRNA–PIWI complexes, the generation of piRNAs is associated with AGO3 or AUB in this procedure. In this cycle, piRNAs form piRNA–AGO3 or piRNA–AUB complexes which provide substrate to each other.44 Ping-pong features have been identified in Drosophila melanogaster, zebra fish, and sponges, but not in mice, indicating that the ping-pong cycle exists in the early stages of evolution.26,45 When piRNAs bind to AGO3, AUB, or PIWI, prominent orientation biases are observed.34,46 In this cycle, AUB-bound piRNAs showed a 5ʹ bias for U, whereas AGO3-bound piRNAs featured an A at position 10 (10A bias). In fly germ cells, AUB associated with a primary piRNA detects and cuts active transposon transcripts through its slicer activity. These cleavage events produce the 5ʹ ends of new piRNAs that are in sense orientation to transposons. Then AGO3–piRNA complexes recognize and cleave cluster transcripts to generate antisense piRNAs after the procedure of loading into AGO3 and maturation through trimming or Zuc.42,43 These phased piRNAs associated with AGO3 or AUB allow target adaptation through sequence diversification.47,48 Studies of PIWI and catalytic inactivation found in various species confirmed the general framework of ping-pong amplification.49,50 A summary of the ping-pong amplification loop of piRNAs is shown in Figure 2.
Mechanisms of action of piRNAs
With the diverse functions of piRNAs, this class of ncRNAs can be classified as master regulators of gene expression. There are several mechanisms underlying piRNA-mediated gene regulation. First, epigenetic mechanisms include DNA methylation and histone modifications. piRNAs play an important role in epigenetic control.51 piRNAs may silence transposons through epigenetic mechanisms to regulate gene expression.52 PIWI is guided by the piRNA to the genomic region, where the piRNA shares complementarities, and this helps in recruiting factors necessary for imparting methylation or acetylation, which are essential for transcriptional regulation of gene expression.15 One example of this mode of gene regulation is the silencing of killer immunoglobulin-like receptors (KIRs).53 The piRNA–PIWI complex can bind to the promoter region and favors recruitment of methylation factors, enabling transcriptional silencing of KIRs. Second, piRNAs also regulate gene expression at the post-transcriptional level via alternative splicing or controlling mRNA stability, and RNA endonucleolytic cleavage or interaction with RNAs.54,55 In general, piRNAs target the transposon sequences found in the 3′-UTRs or 5′-UTRs of mRNAs, and piRNAs derived from pseudogenes and antisense transcripts also target the mRNA of the corresponding endogenous gene, resulting in mRNA deadenylation or degradation.15,56–58 Several studies have confirmed the relationship between piRNAs and their target mRNAs.56,59–61 However, it is still unclear whether the piRNAs have any seed sequences that determine the complementary binding to the target mRNA. Last but not least, piRNAs/piRNA-L have a potential role in regulating gene expression at the translational or post-translational level by directly binding the coding protein in physiological and pathophysiological conditions.62
piRNAs in cancer
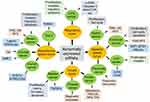
piRNAs have both oncogenic and tumor-suppressive functions in cancer development. Unlike miRNAs, piRNAs are often not complementary to the mRNA of target genes, suggesting that piRNAs play an epigenetic regulatory role in controlling biological functions, including cancer.63,64 Epigenetic alterations, such as DNA hypomethylation and histone hypoacetylation, result in tumor suppressor silencing and oncogenic activation.65–67 In normal cells, a limited set of genes is epigenetically regulated by piRNAs,19,68 allowing the definition of unique tissue signatures based on the profile of piRNAs expressed in given tissues. A study showed that piRNA expression differs significantly across human somatic tissues.69 Variable degrees of heterogeneity in expression patterns are observed in different kinds of tissues. Only a few piRNAs are consistently expressed in both tumor and normal tissues, despite the fact that most of them are encoded in the human genome.69 It has been reported that aberrant expression of piRNAs is a potential cancer-specific signature and can be correlated with clinical features in malignant tissues, indicating an important role for piRNAs in several types of cancers.16 One study69 analyzed more than 6,000 human piRNA transcriptomes and discovered that there were 522 and 273 piRNAs expressed in malignant and tumor-adjacent tissues, respectively. A large body of evidence shows that piRNAs are associated with cancer cell proliferation, differentiation, progression, and metastasis, which could be considered as potential prognostic and diagnostic biomarkers during cancer development. Here, we discuss emerging insights into the roles of piRNAs in several cancers, then describe findings and mechanisms underlying piRNA-mediated gene regulation, and finally point out the potential clinical significance of piRNAs in the management of cancer patients. A summary of the function of piRNAs in different cancer types is shown in Figure 3.
piRNAs in breast cancer
Breast cancer is the most common cancer and the main cause of cancer-related death in women. Aberrant expression of piRNAs is observed in breast cancer and is associated with breast cancer cell proliferation and metastasis, indicating that piRNAs play an important role and are potential biomarkers and therapeutic targets in breast cancer. Evidence also supports the involvement of piRNAs in tumorigenesis via a piRNA-mediated epigenetic mechanism. The highly expressed piR-651 was observed in breast cancer cells.70 A study using deep sequencing to screen out expressed levels of piRNAs in breast cancer and matched non-cancer tissues found that piRNAs, such as piR-20365, piR-20485, piR-20582, and piR-4987, were upregulated in breast cancer tissues, and increased expression of piR-4987 was associated with positive lymph nodes.71 The piRNA biogenesis and effector pathway are present in human breast cancer cells. More than 100 piRNAs were identified to be aberrantly expressed in breast cancer tissues and cells by analyzing small RNA-Seq data, and among them eight piRNAs showed a specific expression pattern in breast cancer target key pathways, suggesting a transcriptional and post-transcriptional regulatory role of piRNAs in cancer.60 Moreover, several studies also found that piRNAs are associated with cell proliferation and metastasis, and with clinical features in patients with breast cancers, indicating that piRNAs may act as biomarkers and therapeutic targets in breast cancer. Another report observed that piR-35127, piR-46545, and piR-52200 could regulate the proliferation of breast cancer cells.72 One report unveiled eight piRNAs as novel independent prognostic markers which were associated with overall survival (OS) by The Cancer Genome Atlas (TCGA) data sets.73 In determining the effects of hormones on piRNA expression in cancer, one study found that the expressions of piR-651 and piR-823 were increased in breast cancer cells after hormone treatment.74 The increased expressions of piR-651 and piR-823 during gonadal development and in cancer may be associated with increased hormone levels and the microenvironment of the cancer.74 In exploring the mechanisms of piRNAs on breast cancer, one of the piRNAs, called piR-932, was highly expressed in breast cancer cells and formed immune complexes through immunoprecipitation with PIWIL2. The combination of piR-932 and PIWIL2 may be a positive regulator in the differentiation of breast cancer stem cells through promoting the methylation of Latexin, a potential putative tumor suppressor and a negative stem cell regulatory gene.75
piRNAs are also believed to play an important role in gene regulation through sequence-specific histone modification and DNA methylation. Genotypic screening of a panel of single-nucleotide polymorphism (SNP)-containing piRNAs found a significant association between SNP rs1326306 G>T in piR-021285 and an increased likelihood of breast cancer. Significant methylation differences were observed at a number of experimentally implicated breast cancer-related genes, and piR-021285 was found to be involved in the regulation of the methylation of the proinvasive Rho GTPase activating protein-11A (ARHGAP11A) gene. This was the first evidence supporting a role for piRNAs in tumorigenesis via a piRNA-mediated epigenetic mechanism.76 A study showed that piR-021285 containing rs1326306 SNP in breast cancer cells reduced methylation at a CpG site within the 5′-UTR/first exon of the ARHGAP11A gene, resulting in augmented migration capability compared to the controls. Furthermore, pi-sno75, a piRNA located in the intronic regions of the growth arrest specific-5 (GAS5) gene, increased the transcription of a pro-apoptotic protein, named tumor necrosis factor (TNF)-related apoptosis inducing ligand (TRAIL), through H3K27 demethylation and H3K4 methylation, and consequently led to the inhibition of breast cancer growth.77 The relationship between piR-36026 and two tumor suppressors, lecithin retinol acyltransferase (LRAT) and serpin family A member-1 (SERPINA1), was also reported, and SERPINA1 and LRAT, the two endogenous piR-36026 target genes, were visualized to interact directly with piR-36026 in MCF7 cells using a piR-36026 molecular beacon assay. Endogenous piR-36026, combined with SERPINA1 and LRAT, could promote cancer progression in MCF7 cells.78 Glycerol-3-phosphate acyltransferase-2 (GPAT2), a member of the testicular cancer gene family, has also been found to be involved in piRNA biogenesis in germline stem cells. Silencing of GPAT2 quantitatively and qualitatively regulated the population of piRNAs, resulting in a more differentiated cancer cell phenotype.79 One report showed that the expression of piR-36712 was significantly lower in breast cancer than that in normal breast tissues. Functional studies found that piR-36712 inhibited expression of SEPW1, a selenium-containing protein, through competition of SEPW1 mRNA with SEPW1P, a retroprocessed pseudogene of SEPW1, RNA for miRNA-7 and miRNA-324. Furthermore, increased piR-36712 had synergistic effects with paclitaxel and doxorubicin in inhibiting proliferation, invasion, and migration in breast cancer cells.55 Overall, piRNAs modulate intracellular signaling to control diverse cellular processes, including proliferation, invasion, migration, apoptosis, and stemness. The above-mentioned studies indicate the involvement of piRNAs in breast cancer. However, the detailed role of piRNAs in breast tumors remains to be thoroughly examined.
piRNAs in lung cancer
Lung cancer is the leading cause of cancer-related death in the world. There are two main groups of lung cancer, namely non-small-cell lung cancer (NSCLC) and small-cell lung cancer (SCLC), with the former comprising nearly 85% of lung cancer cases and the latter approximately 15% of cases. Aberrant piRNA expression is associated with proliferation and metastasis, and may act as a potential diagnostic marker and provide therapeutic targets in lung cancer. piR-55490 could suppress lung cancer growth in vitro and in vivo, and was negatively associated with patient survival.61 piR-L-163 was the most commonly downregulated piRNA in NSCLC compared to the corresponding non-tumor lung tissues,62 while other studies have confirmed the upregulation of piR-651 in lung cancer cells.70–82 In addition, other piRNAs, such as piR-35127 and piR-46545, were downregulated, while piR-34871 and piR-52200 were increased in lung cancer tissues and cells.72 Enforced expression of piR-55490 inhibited cell proliferation, while silencing of piR-55490 led to proliferation being promoted in vitro and in vivo. Moreover, piR-55490 was shown to bind the 3ʹ-UTR of mammalian target of rapamycin (mTOR) mRNA and induce the degradation of mTOR in a mechanism similar to miRNA.61
piRNAs are associated with lung cancer cell proliferation, apoptosis, invasion, metastasis, and chemoresistance to cisplatin (CDDP)-based chemotherapy, and have connections with clinical features in lung cancers. Distinctive piRNA/piRNA-L expression patterns are observed between human bronchial epithelial (HBE) and NSCLC cells. piRNA-like-163 (piR-L-163), the most downregulated piRNA-L in NSCLC cells, binds directly to p-ERM. The piR-L-163/p-ERM interaction is critical for p-ERM’s binding capability to filamentous actin (F-actin) and ERM-binding phosphoprotein-50 (EBP50), which is critical for regulating the migration and invasion of lung cancer cells. Thus, piRNA/piRNA-L may play a regulatory role through direct interaction with proteins in physiological and pathophysiological conditions.62 piR-651 regulates a number of biological functions, as well as carcinogenesis. piR-651 potentially regulates tumorigenic behavior by inhibiting cell proliferation, migration, and invasion, and inducing apoptosis through altering the expressions of the apoptosis-associated proteins cyclin D1 and CDK4 in NSCLC cells.80 Similar observation and conclusion were reported in another study.81 piR-35127, piR-46545, piR-34871, and piR-52200 regulated proliferation and colony formation in normal as well as lung cancer cells, suggesting that these novel piRNAs play a potential role in regulating lung cell transformation and tumorigenesis.72 piRNA-likes (piR-Ls), a novel type of functional sncRNA, play key roles in chemoresistance to CDDP-based chemotherapy in lung squamous cell carcinoma (LSCC). The sensitivity of LSCC to CDDP was enhanced by silencing piR-L-138 both in vivo and in vitro. The increased expression of piR-L-138 was observed in LSCC cells and tumors from patient-derived xenograft models upon CDDP-based chemotherapy. Moreover, piR-L-138 directly interacted with proto-oncogene p60-Murine Double Minute 2 (MDM2) and reversed the CDDP-activated apoptosis in p53-mutated LSCC cells.83 Overall, piRNAs can be used as biomarkers for prognosis, diagnosis, and clinical evaluation, and probably serve as novel therapeutic targets and biomarkers for lung cancer treatment.
piRNAs in gastric cancer
Gastric cancer (GC) is one of the leading causes of cancer-related deaths in the world and is especially prevalent in Asia. Accumulated evidence has elucidated the important roles of piRNAs in GC in recent years. Several reports have investigated the correlation between piRNAs and GC. piR-651 and piR-823 are the most well-understood piRNAs in GC and may be valuable biomarkers with high sensitivity and specificity for detecting GC and distinguishing between different types of GC. Researchers have demonstrated the potential clinical significance of piRNAs in GC.84 The peripheral blood levels of piR-651 and piR-823 in patients with GC were significantly lower than that in healthy controls, and piR-823 was associated with stage and metastasis.84 In addition, increased expression of piR-823 inhibited GC growth both in vitro and in vivo, indicating tumor-suppressive properties.85 However, the opposite findings were reported in another study.70 Thus, the true role of this piRNA remains to be determined. In addition, there are other aberrantly expressed piRNAs in GC, which are associated with clinical features. FR004819 was reported to be positively associated with poor patient survival in GC.19 piR-59056, piR-32105, and piR-58099, which are highly expressed in GC, were involved in the development and recurrence of GC. Moreover, the expression of FR222326, FR290353, FR064000, and FR387750/FR157678 was associated with OS and recurrence-free survival (RFS).86 These findings suggest an important role for piRNAs as promising diagnostic markers and therapeutic targets of GC. Overall, there is limited knowledge about the association between piRNAs and GC biology, particularly the mechanisms underlying this effect. Thus, more effort is warranted to identify more piRNAs and to understand the role of piRNAs in GC biology and their potential clinical applications.
piRNAs in hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the fifth leading cause of cancer death in men and eighth leading cause of death in women in the USA. Several piRNAs have been reported to be dysregulated in HCC and associated with HCC occurrence and development through different signaling pathways and mechanisms, thereby providing new insights into the tumorigenesis of HCC. The upregulated expression of piR-651 has been reported in several cancer types, including HCC.70 It was reported that piR-32299, piR-23670, piR-24684, piR-28488, and piR-7239 were upregulated, while piR-952, piR-820, piR-28525, piR-5938, and piR-5937 were downregulated in HCC tissues. These results suggest that publicly available data can be a useful resource for the identification of sncRNAs, including piRNAs, in HCC progression, and that a data-mining approach is important for biomarker development.87 Combined bioinformatic and biological analyses revealed that piR-Hep1 was highly expressed in HCC compared with the corresponding adjacent non-tumor tissues, and silencing of piR-Hep1 inhibited cell viability, motility, and invasiveness through inactivation of PI3K-Akt signaling, suggesting an oncogenic role for piR-Hep1–PIWIL2 complexes in HCC.88 The expressions of piRNAs, such as Hsa_piR_013306, hsa_piR_00823, hsa_piR_020498, and piR_LLi_30552, were increased in HCC, and functional analysis indicated that these piRNAs can target p53, PI3K/Akt, high mobility group box-1 (HMGB1), PTEN, and tumor necrosis factor (TNF) receptor signaling pathways, which are involved in hepatocarcinogenesis and the development of HCC.89 Overall, the limited results indicate that piRNAs represent a new player in HCC occurrence and development, and a potential prognostic factor that could have diagnostic and therapeutic implications in HCC.
piRNAs in colorectal cancer
Being a major public health burden, colorectal cancer (CRC) is one of the most commonly diagnosed cancers and the major cause of cancer-related death in the world. Aberrant piRNAs are observed in CRC, and are associated with proliferation, cell cycle, and apoptosis, as well as clinical features in CRC. For example, upregulation of piR-651 was confirmed in colon cancer tissues.70 piR-015551 expression was significantly lower in CRC tissues than in normal tissues, and was positively correlated with the lncRNA LNC009643 expression, which included the piR-015551 sequence.90 Large-scale piRNA expression profiling was performed using Illumina® small RNA sequencing in blood serum samples from 403 colon cancer patients and 276 healthy donors. The study found that 31 piRNAs were deregulated in serum of cancer patients compared with healthy donors and that the levels of piR-5937 and piR-28876 may play a role in differentiating between cancer patients and healthy donors, with high sensitivity and specificity.91 piR-hsa-25447, piR-hsa-23992, and piR-hsa-1043 were upregulated, while piR-hsa-28876 was downregulated in CRC tissues.92 piR-019825 contributed independently toward discriminating colon cancer patients from controls.93 In addition, piR-823 was significantly increased in the CRC tissues compared with the adjacent tissues. Inhibition of piR-823 suppressed cell proliferation, cell-cycle arrest, and induced cell apoptosis by targeting heat-shock protein (HSP) 27, 60, and 70 in CRC cells. Moreover, piR-823 increased the transcriptional activity of HSF1, the common transcription factor of HSPs, through binding to HSF1 and activating the phosphorylation at Ser326. This highlighted the potential therapeutic target of piR-823 in CRC.94 Based upon piRNA:mRNA sequence complementarities, a panel of tumor suppressor genes was identified as direct targets of piR-1245, and an inverse correlation between their expression and piR-1245 in CRC was observed. This identified the role of piR-1245 as a novel oncogene and a potential prognostic biomarker in CRC.95 Overall, the precise mechanisms of the interaction between piRNAs and relevant targets merit further investigation. The studies summarized herein have identified the different roles for a piRNA acting as a novel oncogene or tumor suppressor, and a potential prognostic biomarker in CRC. With the help of available tumor samples with extensive clinical annotations and improved profiling platforms, more novel CRC-related piRNAs could be identified, which will further update our knowledge on the mechanistic and prognostic roles of piRNAs in CRC.
piRNAs in renal cell carcinoma
Renal cell carcinoma (RCC) is a major human malignancy. About 30% of newly diagnosed cases have evidence of metastases at diagnosis. Understanding the molecular mechanisms of RCC metastasis and finding biomarkers for early prediction may help to guide more appropriate treatment and to develop target-specific molecular therapies. Aberrant piRNA expression is observed in RCC, and is associated with proliferation and metastasis in RCC. By deep sequencing 24 frozen benign kidney and clear-cell renal cell carcinoma (ccRCC) specimens, and using the publicly available piRNA database, 19 piRNAs were found to be differentially expressed in these 24 specimens, and upregulation of piR-32051, piR-39894, and piR-43607 was highly associated with ccRCC metastasis, clinical stage, and poor cancer-specific survival.96 piR-823 expression was downregulated in tumor specimens using a qRT-PCR assay, and was found to be positively correlated with worse outcome, indicating its complex role in RCC pathogenesis.97 Researchers identified 235 upregulated and 369 downregulated piRNAs from 106 ccRCC patient samples by piRNA microarray, and found that piR-38756 and piR-30924 were upregulated, while piR-57125 was downregulated in metastatic primary tumors and bone metastases compared with non-metastatic primary tumors.98 piR-32051, piR-39894, and piR-43607 were found to be derived from a piRNA cluster at chromosome 17 and belonged to metastasis-related piRNAs, which were highly associated with tumor stage, metastasis, and short patient survival in a 68-case formalin-fixed and paraffin-embedded ccRCC tissue cohort.96 Expression levels of piR-823 were deregulated in tumor tissues, blood serum, and urine of patients with RCC. There was a significant reduction of piR-823 in tumor tissues, while the expression of piR-823 was significantly higher in blood serum and urine in patients with RCC compared to healthy individuals. Higher levels of piR-823 in tumor tissues were associated with shorter disease-free survival of patients; however, unsatisfactory analytical performance was observed in blood serum samples. Thus, the complex role of piRNAs in RCC pathogenesis still needs to be elucidated.97 The higher expression of piR-30924 and piR-38756, and the lower expression of piR-57125, in metastatic primary ccRCC were significantly associated with recurrence and OS. Multivariate Cox regression analyses showed that piR-30924 and piR-57125 acted as independent prognostic biomarkers in combination with clinical and pathological factors.98 Further study of piRNAs may help researchers to understand the mechanisms of RCC metastasis and progression.
piRNAs in head and neck squamous cell carcinoma
Head and neck squamous cell carcinoma (HNSCC) represents the sixth most deadly cancer worldwide. Despite advances in diagnosis and treatment, the mortality of patients with HNSCC has improved minimally in recent years. Aberrant expression of piRNAs is observed in HNSCC. The specific piRNAs have connection with clinical features and the piRNA expression patterns are associated with human papillomavirus (HPV) status and HPV type, indicating the potential value of piRNA in assessing HNSCC patient prognosis. Analysis of piRNA levels in HNSCC and non-malignant tissues revealed distinct expression patterns. The expression of specific piRNAs is deregulated in HNSCC, and changes with both HPV status and type. Importantly, five piRNAs, FR018916, FR140858, FR197104, FR237180, and FR298757, in which expression in HPV-positive HNSCC tumors was associated with worse OS, highlighted the potential utility of piRNAs in patient management.99 Similarly, an expression pattern for a panel of 41 piRNAs could be used to differentiate between HPV-positive and HPV-negative HNSCC samples, with 11 of them being overexpressed distinctively in HPV-16- or HPV-18-induced tumors. Among them, piR-35953, piR-36984, piR-39592, piR-36715, and piR-305065 were associated with OS.100 NONHSAT123636 and NONHSAT113708 were reported to be directly associated with tumor stage, and NONHSAT067200 in smoking-related HNSCC was linked to patient survival.101 Researchers identified a panel of 30 HPV-dysregulated piRNAs between HPV-16-positive HNSCC and HPV-negative normal samples from RNA-sequencing data sets from TCGA, and NONHSAT077364, NONHSAT102574, and NONHSAT128479 were later verified in vitro in HPV-positive cancer cells and HPV-negative normal cells, indicating a potential role for these piRNAs in the pathogenesis and progression of HNSCC.102
Researchers have also explored the mechanisms of action of piRNAs on HNSCC. One study found that piR-34736 correlated with PRDM9 mutation and decreased Bax/Bcl2 ratio, and induced vimentinin expression in UMSCC-10B and HN-30 cells.103 Others found that several piRNAs altered PIWIL1 protein expression and correlated with genomic alterations common to smoking-related HNSCC, such as TP53 mutation, TP53-3p co-occurrence, and 3q26, 8q24, and 11q13 amplification. This provided novel insights into the etiology-specific piRNA landscape of smoking-induced HNSC.101 Overall, rigorous studies on piRNA are lacking, with limited knowledge on their functionality in HPV-induced HNSCC or mechanistic involvement in malignant transformation. The expression of specific piRNAs is deregulated in HNSCC, and changes with HPV status and HPV type prompt investigation of piRNA dysregulation in HPV-related HNSCC. This highlights the potential value of piRNAs in patient management, introduces new insight into their role in the pathogenesis of HNSCC, and implicates a novel panel of piRNAs with potential utility as prognostic markers and therapeutic targets.
piRNAs in other cancer types
The expression of piR-651 was found to be increased in cervical cancer cells.70 FR090905 improved overall survival, while FR027884 was associated with poor patient outcome in cervical cancer, as reported by researchers via analysis of more than 6,000 human piRNA transcriptomes derived from cervical cancer tissues and non-malignant tissue.19 Following the overexpression of HILI, the retrotransposon elements LINE1 and LINE1-associated small RNAs were decreased in HeLa cells.104 piR-49322 was observed to locate in the nucleolus and around the periphery of the nuclear membrane in HeLa cells using an in situ hybridization assay, which was consistent with the report that mature piRNAs were generated in a germline-specific perinuclear structure and then imported into the nucleus in the form of PIWI–piRNA complexes.105 piR-017061 was dramatically reduced in pancreatic ductal adenocarcinoma compared to tissues from normal pancreas.106 piR-52207 and piR-33733 were upregulated in ovarian cancer, and modulated processes and key signaling pathways in ovarian oncogenesis.107 piRNA DQ594040 was the most downregulated piRNA and overexpression of DQ594040 inhibited proliferation and colony formation, and promoted cell apoptosis by increasing the expression of tumor necrosis factor superfamily-4 (TNFSF4) in bladder cancer cells. About 106 piRNAs were upregulated, while 91 were downregulated in three bladder cancer tissues via piRNA microarray assays used to investigate global piRNA expression.108 Among them, piRABC was the most upregulated piRNA, which regulated cell proliferation and colony formation, and promoted cell apoptosis in bladder cancer cells through TNFSF4. piR-823 acted in an oncogenic role and was upregulated in multiple myeloma (MM) patients and MM cells, and correlated with poor prognosis. piR-823 could promote MM by directly regulating DNA methyltransferases 3A and 3B (DNMT3A, DNMT3B) to increase global methylation. piR-598 impacted cell survival and promoted cell proliferation in glioma.109 It was reported that PIWIL1–piRNA-DQ593109 complexes were overexpressed in glioma endothelial cells, and increased blood–tumor barrier permeability by regulating the lncRNA maternally expressed-3 (MEG3)/miR-330-5p/runt-related transcription factor-3 (RUNX3) regulatory loop.110 In leukemia cells, overexpression of CDKN2B-related piRNA hsa_piR_011186 promoted cell-cycle progression and inhibited apoptosis.111 These effects were probably mediated by the piRNA complex of cyclin-dependent kinase inhibitor-2B (CDKN2B)-related piRNA that were associated with DNMT1 and/or enhancer of zeste homolog-2 (EZH2) to modulate the methylation of DNA and H3 in the promoter region of the CDKN2B gene, highlighting epigenetic modifications on the promoter of cell-cycle regulating gene.111 Several piRNAs were aberrantly expressed in testicular germ cell tumors (TGCTs). Reduced expression of the piRNAs DQ598918, DQ589977, and DQ601609 was observed in both seminoma and non-seminoma tumors.112 piR-004172, piR-006113, and piR-007509 were shown to be downregulated in TGCT compared to the normal testis by small RNA sequencing.113 These results facilitate our understanding of the functions and roles of piRNAs in cancer cell proliferation, metastasis, and invasion. Also, as biomarkers for clinical application, piRNAs have the advantage of short size, enabling them to pass through cell membranes easily without being degraded like long RNAs, and are easily detectable in serum and other samples of patients. It have been reported that piRNAs in these samples from patients with CRC and other cancers were dysregulated.93 Therefore, further effort in this area is expected to improve our understanding of the diagnosis and management of human cancers including CRC.
Therapeutic insights into piRNAs
Beyond the role of piRNAs as biomarkers, studies have also demonstrated their potential role as therapeutic tools. It has been reported that artificial piRNAs, generated through the expression of sense and antisense transcripts, were sufficient to induce epigenetic silencing of the target gene in a mouse model,114 highlighting the potential of piRNAs as clinical therapeutic targets. According to the characteristics of piRNAs, several therapeutic strategies could be designed. The most appealing strategy involves using synthetic piRNAs, which could block the synthesis of cancer-related proteins by binding to mRNAs like miRNAs. This has the advantage of not requiring processing by enzymes, such as Dicer, required by miRNAs.115 Another speculative advantage of piRNAs over miRNAs is the possibility of targets with better specificity, since each miRNA regulates several mRNAs.115 piRNAs may directly regulate the expression of PIWI. Studies demonstrated that inhibition of PIWI proteins could inhibit cell proliferation and the cell cycle, and induce apoptosis, and PIWI proteins were negatively associated with patient OS.116,117 Moreover, PIWI can increase resistance to chemotherapy such as cisplatin.118,119 Thus, an alternative strategy for treatment based on piRNAs is to design specific piRNAs to bind PIWI proteins; in this way, piRNAs could silence PIWI genes at the transcriptional level and block the harmful outcomes. Silencing PIWI proteins and piRNAs acted as oncogenes in cancer or overexpression of piRNAs acted as tumor suppressors can alter the recruitment of epigenetic factors, thereby influencing the progression of cancer. It is becoming clear that piRNAs are involved in tumorigenesis and tumor development. Dysregulation of piRNAs is involved in cancer cell proliferation, invasiveness, and malignant cell transformation through transcriptional, post-transcriptional, and epigenetic regulatory mechanisms. Modulating the expression of piRNAs may reverse these phenotypes. While the exact mechanism remains to be determined, it is likely that piRNAs interact with mRNAs through complementary sequences, leading to degradation or inhibition of translation of mRNA. While investigations into therapeutic methods based on piRNAs are still in the early stages, more therapeutic achievements could be reached in the future with an increased understanding of the mechanisms and functions of piRNAs in cancer.
Conclusion and future perspectives
piRNAs are gaining increased attention with the development of high-throughput sequencing and bioinformatic technologies. In particular, piRNAs have emerged as a highly promising area of study given their gene-regulatory function in both the nucleus and the cytoplasm. While there is still incomplete understanding of the functions and interactions of piRNAs and PIWI, the biogenesis and functions of piRNAs need further elucidation to improve our understanding of their role in the development of diverse diseases, especially in cancers. At present, piRNA has been recognized to express aberrantly in a cancer-specific manner across a variety of tumor types, although the oncogenic or tumor-suppressing mechanisms of piRNAs still need to be explored. It is also crucial to explore the underlying molecular mechanism by which dysregulation of piRNAs is involved in tumorigenesis. Although numerous studies have demonstrated that piRNAs are dysregulated in cancers and may serve as a novel diagnostic biomarker and therapeutic target for cancer, challenges exist which hinder the immediate clinical application of piRNAs. First, the choice of samples for comparison with tumor tissues is controversial and debatable. Some researchers preferred the use of tumor-adjacent normal tissues, as they were scored for the presence of tumor cells,120,121 while others insisted that there was a neighboring effect of tumors on these tissues in cancers such as prostate and breast cancer.122–124 One study125 found that a number of genes were activated in the tumor-adjacent mucosa similarly to the tumor tissues in CRC, while they were inactivated in normal tissues, indicating large differences between the tumor-adjacent and normal tissues. Second, handling of large amounts of data is difficult and challenging. However, a large number of piRNAs may be rare or expressed at a low level, and a single count could result in technical artifacts. Thus, The use of piRNAs as therapeutic tools also raises challenges. Nevertheless, understanding of piRNAs' complex interactions will provide many novel interventions in both medical discovery and clinical practice. In the future, more piRNAs will be identified and the mechanistic role of piRNAs in tumor biology is expected to provide novel insights into their role in cancer, bringing them into the clinical arena for diagnosis, prognosis, and therapeutics in cancer patients.
Acknowledgments
This work was supported in part by grants from the National Nature Scientific Foundation of China (Nos. 81473716, 81871863) and the Major Program of National Natural Science Foundation of Guangdong (No. 2018B030311061).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi:10.1038/nature11233
2. Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319(5871):1787–1789. doi:10.1126/science.1155472
3. Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi:10.1038/nature11247
4. Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochim Biophys Acta. 2014;1839(11):1097–1109. doi:10.1016/j.bbagrm.2014.08.012
5. Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–149. doi:10.1038/nrg2904
6. Iyengar BR, Choudhary A, Sarangdhar MA, Venkatesh KV, Gadgil CJ, Pillai B. Non-coding RNA interact to regulate neuronal development and function. Front Cell Neurosci. 2014;8:47. doi:10.3389/fncel.2014.00052
7. Place RF, Noonan EJ. Non-coding RNAs turn up the heat: an emerging layer of novel regulators in the mammalian heat shock response. Cell Stress Chaperones. 2014;19(2):159–172. doi:10.1007/s12192-013-0456-5
8. Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev. 2004;18(17):2068–2073. doi:10.1101/gad.1219904
9. Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi:10.1126/science.1138341
10. Taft RJ, Glazov EA, Cloonan N, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41(5):572–578. doi:10.1038/ng.312
11. Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442(7099):199–202. doi:10.1038/nature04917
12. Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313(5785):320–324. doi:10.1126/science.1129333
13. Peng JC, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr Opin Cell Biol. 2013;25(2):190–194. doi:10.1016/j.ceb.2013.01.010
14. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–D73. doi:10.1093/nar/gkt1181
15. Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505(7483):353–359. doi:10.1038/nature12987
16. Siddiqi S, Matushansky I. Piwis and piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem. 2012;113(2):373–380. doi:10.1002/jcb.23363
17. Weng W, Li H, Goel A. Piwi-interacting RNAs (piRNAs) and cancer: emerging biological concepts and potential clinical implications. Biochim Biophys Acta Rev Cancer. 2019;1871(1):160–169. doi:10.1016/j.bbcan.2018.12.005
18. Assumpcao CB, Calcagno DQ, Araujo TM, et al. The role of piRNA and its potential clinical implications in cancer. Epigenomics. 2015;7(6):975–984. doi:10.2217/epi.15.37
19. Martinez VD, Vucic EA, Thu KL, et al. Unique somatic and malignant expression patterns implicate PIWI-interacting RNAs in cancer-type specific biology. Sci Rep. 2015;5:10423. doi:10.1038/srep10423
20. Zhuang K, Wu Q, Jin CS, Yuan HJ, Cheng JZ. Long non-coding RNA HNF1A-AS is upregulated and promotes cell proliferation and metastasis in nasopharyngeal carcinoma. Cancer Biomark. 2016;16(2):291–300. doi:10.3233/CBM-150567
21. Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH, Li W. PIWI proteins and PIWI-interacting RNA: emerging roles in cancer. Cell Physiol Biochem. 2017;44(1):1–20.
22. Krishnan P, Damaraju S. The challenges and opportunities in the clinical application of noncoding RNAs: the road map for miRNAs and piRNAs in cancer diagnostics and prognostics. Int J Genomics. 2018;2018:5848046. doi:10.1155/2018/5848046
23. Li J, Shen H, Xie H, et al. Dysregulation of ncRNAs located at the DLK1DIO3 imprinted domain: involvement in urological cancers. Cancer Manag Res. 2019;11:777–787. doi:10.2147/CMAR.S190764
24. Grivna ST, Beyret E, Wang Z, Lin H. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 2006;20(13):1709–1714. doi:10.1101/gad.1434406
25. Watanabe T, Takeda A, Tsukiyama T, et al. Identification and characterization of two novel classes of small RNAs in the mouse germline: retrotransposon-derived siRNAs in oocytes and germline small RNAs in testes. Genes Dev. 2006;20(13):1732–1743. doi:10.1101/gad.1425706
26. Williams Z, Morozov P, Mihailovic A, et al. Discovery and characterization of piRNAs in the human fetal ovary. Cell Rep. 2015;13(4):854–863. doi:10.1016/j.celrep.2015.09.030
27. Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi:10.1146/annurev.cellbio.24.110707.175327
28. Weick EM, Miska EA. piRNAs: from biogenesis to function. Development. 2014;141(18):3458–3471. doi:10.1242/dev.094037
29. Ha H, Song J, Wang S, et al. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genomics. 2014;15:545. doi:10.1186/1471-2164-15-545
30. Krishnan P, Ghosh S, Wang B, et al. Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci Rep. 2016;6:32843. doi:10.1038/srep32843
31. Keam SP, Young PE, McCorkindale AL, et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res. 2014;42(14):8984–8995. doi:10.1093/nar/gku620
32. Zhong F, Zhou N, Wu K, et al. A SnoRNA-derived piRNA interacts with human interleukin-4 pre-mRNA and induces its decay in nuclear exosomes. Nucleic Acids Res. 2015;43(21):10474–10491. doi:10.1093/nar/gkv954
33. Cox DN, Chao A, Lin H. piwi encodes a nucleoplasmic factor whose activity modulates the number and division rate of germline stem cells. Development. 2000;127(3):503–514.
34. Brennecke J, Aravin AA, Stark A, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128(6):1089–1103. doi:10.1016/j.cell.2007.01.043
35. Grishok A, Tabara H, Mello CC. Genetic requirements for inheritance of RNAi in C. elegans. Science. 2000;287(5462):2494–2497.
36. Pak J, Fire A. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007;315(5809):241–244. doi:10.1126/science.1132839
37. Luteijn MJ, Ketting RF. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 2013;14(8):523–534. doi:10.1038/nrg3495
38. Zamore PD. Somatic piRNA biogenesis. Embo J. 2010;29(19):3219–3221. doi:10.1038/emboj.2010.232
39. Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128(4):669–681. doi:10.1016/j.cell.2007.01.033
40. Han BW, Zamore PD. piRNAs. Curr Biol. 2014;24(16):R730–R733. doi:10.1016/j.cub.2014.07.037
41. Vourekas A, Zheng Q, Alexiou P, et al. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 2012;19(8):773–781. doi:10.1038/nsmb.2347
42. Mohn F, Handler D, Brennecke J. Noncoding RNA. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science. 2015;348(6236):812–817. doi:10.1126/science.aaa1039
43. Han BW, Wang W, Li C, Weng Z, Zamore PD. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science. 2015;348(6236):817–821. doi:10.1126/science.aaa1264
44. Grimson A, Srivastava M, Fahey B, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455(7217):1193–1197. doi:10.1038/nature07415
45. Beyret E, Liu N, Lin H. piRNA biogenesis during adult spermatogenesis in mice is independent of the ping-pong mechanism. Cell Res. 2012;22(10):1429–1439. doi:10.1038/cr.2012.120
46. Gunawardane LS, Saito K, Nishida KM, et al. A slicer-mediated mechanism for repeat-associated siRNA 5ʹ end formation in Drosophila. Science. 2007;315(5818):1587–1590. doi:10.1126/science.1140494
47. Wang W, Han BW, Tipping C, et al. Slicing and binding by Ago3 or Aub trigger piwi-bound piRNA production by distinct mechanisms. Mol Cell. 2015;59(5):819–830. doi:10.1016/j.molcel.2015.08.007
48. Senti KA, Jurczak D, Sachidanandam R, Brennecke J. piRNA-guided slicing of transposon transcripts enforces their transcriptional silencing via specifying the nuclear piRNA repertoire. Genes Dev. 2015;29(16):1747–1762. doi:10.1101/gad.267252.115
49. De Fazio S, Bartonicek N, Di Giacomo M, et al. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature. 2011;480(7376):259–263. doi:10.1038/nature10547
50. Wang W, Yoshikawa M, Han BW, et al. The initial uridine of primary piRNAs does not create the tenth adenine that Is the hallmark of secondary piRNAs. Mol Cell. 2014;56(5):708–716. doi:10.1016/j.molcel.2014.10.016
51. Wei JW, Huang K, Yang C, Kang CS. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9. doi:10.3892/or.2016.5236
52. Moyano M, Stefani G. piRNA involvement in genome stability and human cancer. J Hematol Oncol. 2015;8:38. doi:10.1186/s13045-015-0133-5
53. Cichocki F, Lenvik T, Sharma N, Yun G, Anderson SK, Miller JS. Cutting edge: KIR antisense transcripts are processed into a 28-base PIWI-like RNA in human NK cells. J Immunol. 2010;185(4):2009–2012. doi:10.4049/jimmunol.1000855
54. Gu K, Mok L, Chong MMW. Regulating gene expression in animals through RNA endonucleolytic cleavage. Heliyon. 2018;4(11):e00908. doi:10.1016/j.heliyon.2018.e00908
55. Tan L, Mai D, Zhang B, et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer. 2019;18(1):9. doi:10.1186/s12943-019-1010-6
56. Watanabe T, Lin H. Posttranscriptional regulation of gene expression by Piwi proteins and piRNAs. Mol Cell. 2014;56(1):18–27. doi:10.1016/j.molcel.2014.09.012
57. Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12(4):246–258. doi:10.1038/nrm3089
58. Balaratnam S, West N, Basu S. A piRNA utilizes HILI and HIWI2 mediated pathway to down-regulate ferritin heavy chain 1 mRNA in human somatic cells. Nucleic Acids Res. 2018;46(20):10635–10648. doi:10.1093/nar/gky728
59. Chu H, Hui G, Yuan L, et al. Identification of novel piRNAs in bladder cancer. Cancer Lett. 2015;356(2 Pt B):561–567. doi:10.1016/j.canlet.2014.10.004
60. Hashim A, Rizzo F, Marchese G, et al. RNA sequencing identifies specific PIWI-interacting small non-coding RNA expression patterns in breast cancer. Oncotarget. 2014;5(20):9901–9910. doi:10.18632/oncotarget.2476
61. Peng L, Song L, Liu C, et al. piR-55490 inhibits the growth of lung carcinoma by suppressing mTOR signaling. Tumour Biol. 2016;37(2):2749–2756. doi:10.1007/s13277-015-4056-0
62. Mei Y, Wang Y, Kumari P, et al. A piRNA-like small RNA interacts with and modulates p-ERM proteins in human somatic cells. Nat Commun. 2015;6:7316. doi:10.1038/ncomms8316
63. Wang QX, Zhu YQ, Zhang H, Xiao J. Altered MiRNA expression in gastric cancer: a systematic review and meta-analysis. Cell Physiol Biochem. 2015;35(3):933–944. doi:10.1159/000369750
64. Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8(2):188–194. doi:10.1038/ncb1353
65. Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775(1):138–162. doi:10.1016/j.bbcan.2006.08.007
66. Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005;2 Suppl 1(Suppl 1):S4–S11. doi:10.1038/ncponc0354
67. Guo Z, Maki M, Ding R, Yang Y, Zhang B, Xiong L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci Rep. 2014;4:5150. doi:10.1038/srep05150
68. Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8(4):286–298. doi:10.1038/nrg2005
69. Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi:10.1038/nrc3130
70. Cheng J, Guo JM, Xiao BX, et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412(17–18):1621–1625. doi:10.1016/j.cca.2011.05.015
71. Huang G, Hu H, Xue X, et al. Altered expression of piRNAs and their relation with clinicopathologic features of breast cancer. Clin Transl Oncol. 2013;15(7):563–568. doi:10.1007/s12094-012-0966-0
72. Reeves ME, Firek M, Jliedi A, Amaar YG. Identification and characterization of RASSF1C piRNA target genes in lung cancer cells. Oncotarget. 2017;8(21):34268–34282. doi:10.18632/oncotarget.15965
73. Krishnan P, Ghosh S, Graham K, Mackey JR, Kovalchuk O, Damaraju S. Piwi-interacting RNAs and PIWI genes as novel prognostic markers for breast cancer. Oncotarget. 2016;7(25):37944–37956. doi:10.18632/oncotarget.9272
74. Oner C, Turgut Cosan D, Colak E. Estrogen and androgen hormone levels modulate the expression of Piwi interacting rna in prostate and breast cancer. PLoS One. 2016;11(7):e0159044. doi:10.1371/journal.pone.0159044
75. Zhang H, Ren Y, Xu H, Pang D, Duan C, Liu C. The expression of stem cell protein Piwil2 and piR-932 in breast cancer. Surg Oncol. 2013;22(4):217–223. doi:10.1016/j.suronc.2013.07.001
76. Fu A, Jacobs DI, Hoffman AE, Zheng T, Zhu Y. PIWI-interacting RNA 021285 is involved in breast tumorigenesis possibly by remodeling the cancer epigenome. Carcinogenesis. 2015;36(10):1094–1102. doi:10.1093/carcin/bgv105
77. He X, Chen X, Zhang X, et al. An Lnc RNA (GAS5)/SnoRNA-derived piRNA induces activation of TRAIL gene by site-specifically recruiting MLL/COMPASS-like complexes. Nucleic Acids Res. 2015;43(7):3712–3725. doi:10.1093/nar/gkv214
78. Lee YJ, Moon SU, Park MG, et al. Multiplex bioimaging of piRNA molecular pathway-regulated theragnostic effects in a single breast cancer cell using a piRNA molecular beacon. Biomaterials. 2016;101:143–155. doi:10.1016/j.biomaterials.2016.05.052
79. Lacunza E, Montanaro MA, Salvati A, et al. Small non-coding RNA landscape is modified by GPAT2 silencing in MDA-MB-231 cells. Oncotarget. 2018;9(46):28141–28154. doi:10.18632/oncotarget.25582
80. Zhang SJ, Yao J, Shen BZ, et al. Role of piwi-interacting RNA-651 in the carcinogenesis of non-small cell lung cancer. Oncol Lett. 2018;15(1):940–946. doi:10.3892/ol.2017.7406
81. Yao J, Wang YW, Fang BB, Zhang SJ, Cheng BL. piR-651 and its function in 95-D lung cancer cells. Biomed Rep. 2016;4(5):546–550. doi:10.3892/br.2016.628
82. Li D, Luo Y, Gao Y, et al. piR-651 promotes tumor formation in non-small cell lung carcinoma through the upregulation of cyclin D1 and CDK4. Int J Mol Med. 2016;38(3):927–936. doi:10.3892/ijmm.2016.2671
83. Wang Y, Gable T, Ma MZ, et al. A piRNA-like small RNA induces chemoresistance to cisplatin-based therapy by inhibiting apoptosis in lung squamous cell carcinoma. Mol Ther Nucleic Acids. 2017;6:269–278. doi:10.1016/j.omtn.2017.01.003
84. Cui L, Lou Y, Zhang X, et al. Detection of circulating tumor cells in peripheral blood from patients with gastric cancer using piRNAs as markers. Clin Biochem. 2011;44(13):1050–1057. doi:10.1016/j.clinbiochem.2011.06.004
85. Cheng J, Deng H, Xiao B, et al. piR-823, a novel non-coding small RNA, demonstrates in vitro and in vivo tumor suppressive activity in human gastric cancer cells. Cancer Lett. 2012;315(1):12–17. doi:10.1016/j.canlet.2011.10.004
86. Martinez VD, Enfield KS, Rowbotham DA, Lam WL. An atlas of gastric PIWI-interacting RNA transcriptomes and their utility for identifying signatures of gastric cancer recurrence. Gastric Cancer. 2016;19(2):660–665. doi:10.1007/s10120-015-0487-y
87. Koduru SV, Leberfinger AN, Kawasawa YI, et al. Non-coding RNAs in various stages of liver disease leading to hepatocellular carcinoma: differential expression of miRNAs, piRNAs, lncRNAs, circRNAs, and sno/mt-RNAs. Sci Rep. 2018;8(1):7967. doi:10.1038/s41598-018-26360-1
88. Law PT, Qin H, Ching AK, et al. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58(6):1165–1173. doi:10.1016/j.jhep.2013.01.032
89. Rizzo F, Rinaldi A, Marchese G, et al. Specific patterns of PIWI-interacting small noncoding RNA expression in dysplastic liver nodules and hepatocellular carcinoma. Oncotarget. 2016;7(34):54650–54661. doi:10.18632/oncotarget.10567
90. Chu H, Xia L, Qiu X, et al. Genetic variants in noncoding PIWI-interacting RNA and colorectal cancer risk. Cancer. 2015;121(12):2044–2052. doi:10.1002/cncr.29314
91. Vychytilova-Faltejskova P, Stitkovcova K, Radova L, et al. Circulating PIWI-interacting RNAs piR-5937 and piR-28876 are promising diagnostic biomarkers of colon cancer. Cancer Epidemiol Biomarkers Prev. 2018;27(9):1019–1028. doi:10.1158/1055-9965.EPI-18-0318
92. Koduru SV, Tiwari AK, Hazard SW, Mahajan M, Ravnic DJ. Exploration of small RNA-seq data for small non-coding RNAs in human colorectal cancer. J Genomics. 2017;5:16–31. doi:10.7150/jgen.18856
93. Yuan T, Huang X, Woodcock M, et al. Plasma extracellular RNA profiles in healthy and cancer patients. Sci Rep. 2016;6:19413. doi:10.1038/srep19413
94. Yin J, Jiang XY, Qi W, et al. piR-823 contributes to colorectal tumorigenesis by enhancing the transcriptional activity of HSF1. Cancer Sci. 2017;108(9):1746–1756. doi:10.1111/cas.13300
95. Weng W, Liu N, Toiyama Y, et al. Novel evidence for a PIWI-interacting RNA (piRNA) as an oncogenic mediator of disease progression, and a potential prognostic biomarker in colorectal cancer. Mol Cancer. 2018;17(1):16. doi:10.1186/s12943-018-0767-3
96. Li Y, Wu X, Gao H, et al. Piwi-Interacting RNAs (piRNAs) are dysregulated in renal cell carcinoma and associated with tumor metastasis and cancer-specific survival. Mol Med. 2015;21:381–388. doi:10.2119/molmed.2014.00203
97. Iliev R, Fedorko M, Machackova T, et al. Expression levels of PIWI-interacting RNA, piR-823, are deregulated in tumor tissue, blood serum and urine of patients with renal cell carcinoma. Anticancer Res. 2016;36(12):6419–6423. doi:10.21873/anticanres.11239
98. Busch J, Ralla B, Jung M, et al. Piwi-interacting RNAs as novel prognostic markers in clear cell renal cell carcinomas. J Exp Clin Cancer Res. 2015;34:61. doi:10.1186/s13046-015-0180-3
99. Firmino N, Martinez VD, Rowbotham DA, Enfield KSS, Bennewith KL, Lam WL. HPV status is associated with altered PIWI-interacting RNA expression pattern in head and neck cancer. Oral Oncol. 2016;55:43–48. doi:10.1016/j.oraloncology.2016.01.012
100. Irimie AI, Braicu C, Sonea L, et al. A looking-glass of non-coding RNAs in oral cancer. Int J Mol Sci. 2017;18(12):
101. Krishnan AR, Korrapati A, Zou AE, et al. Smoking status regulates a novel panel of PIWI-interacting RNAs in head and neck squamous cell carcinoma. Oral Oncol. 2017;65:68–75. doi:10.1016/j.oraloncology.2016.12.022
102. Krishnan AR, Qu Y, Li PX, et al. Computational methods reveal novel functionalities of PIWI-interacting RNAs in human papillomavirus-induced head and neck squamous cell carcinoma. Oncotarget. 2018;9(4):4614–4624. doi:10.18632/oncotarget.23464
103. Zou AE, Zheng H, Saad MA, et al. The non-coding landscape of head and neck squamous cell carcinoma. Oncotarget. 2016;7(32):51211–51222. doi:10.18632/oncotarget.9979
104. Lu Y, Li C, Zhang K, et al. Identification of piRNAs in Hela cells by massive parallel sequencing. BMB Rep. 2010;43(9):635–641. doi:10.5483/BMBRep.2010.43.9.635
105. Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Development. 2008;135(1):3–9. doi:10.1242/dev.006486
106. Muller S, Raulefs S, Bruns P, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi:10.1186/s12943-014-0278-9
107. Singh G, Roy J, Rout P, Mallick B. Genome-wide profiling of the PIWI-interacting RNA-mRNA regulatory networks in epithelial ovarian cancers. PLoS One. 2018;13(1):e0190485. doi:10.1371/journal.pone.0190485
108. Yan H, Wu QL, Sun CY, et al. piRNA-823 contributes to tumorigenesis by regulating de novo DNA methylation and angiogenesis in multiple myeloma. Leukemia. 2015;29(1):196–206. doi:10.1038/leu.2014.135
109. Jacobs DI, Qin Q, Lerro MC, et al. PIWI-interacting RNAs in gliomagenesis: evidence from post-GWAS and functional analyses. Cancer Epidemiol Biomarkers Prev. 2016;25(7):1073–1080. doi:10.1158/1055-9965.EPI-16-0047
110. Shen S, Yu H, Liu X, et al. PIWIL1/piRNA-DQ593109 regulates the permeability of the blood-tumor barrier via the MEG3/miR-330-5p/RUNX3 axis. Mol Ther Nucleic Acids. 2018;10:412–425. doi:10.1016/j.omtn.2017.12.020
111. Wu D, Fu H, Zhou H, Su J, Zhang F, Shen J. Effects of novel ncRNA molecules, p15-piRNAs, on the methylation of DNA and histone H3 of the CDKN2B promoter region in U937 cells. J Cell Biochem. 2015;116(12):2744–2754. doi:10.1002/jcb.25199
112. Ferreira HJ, Heyn H. Garcia del Muro X, et al. Epigenetic loss of the PIWI/piRNA machinery in human testicular tumorigenesis. Epigenetics. 2014;9(1):113–118. doi:10.4161/epi.27237
113. Rounge TB, Furu K, Skotheim RI, Haugen TB, Grotmol T, Enerly E. Profiling of the small RNA populations in human testicular germ cell tumors shows global loss of piRNAs. Mol Cancer. 2015;14:153. doi:10.1186/s12943-014-0278-9
114. Itou D, Shiromoto Y, Yukiho SY, et al. Induction of DNA methylation by artificial piRNA production in male germ cells. Curr Biol. 2015;25(7):901–906. doi:10.1016/j.cub.2015.01.060
115. Xie T, Huang M, Wang Y, Wang L, Chen C, Chu X. MicroRNAs as regulators, biomarkers and therapeutic targets in the drug resistance of colorectal cancer. Cell Physiol Biochem. 2016;40(1–2):62–76. doi:10.1159/000452525
116. Lu Y, Zhang K, Li C, et al. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One. 2012;7(1):e30999. doi:10.1371/journal.pone.0030999
117. Li D, Sun X, Yan D, et al. Piwil2 modulates the proliferation and metastasis of colon cancer via regulation of matrix metallopeptidase 9 transcriptional activity. Exp Biol Med (Maywood). 2012;237(10):1231–1240. doi:10.1258/ebm.2012.011380
118. Yuan Y, Wang H, Wu Y, et al. P53 contributes to cisplatin induced renal oxidative damage via regulating P66shc and MnSOD. Cell Physiol Biochem. 2015;37(4):1240–1256. doi:10.1159/000430247
119. Wang QE, Han C, Milum K, Wani AA. Stem cell protein Piwil2 modulates chromatin modifications upon cisplatin treatment. Mutat Res. 2011;708(1–2):59–68. doi:10.1016/j.mrfmmm.2011.02.001
120. Okugawa Y, Toiyama Y, Toden S, et al. Clinical significance of SNORA42 as an oncogene and a prognostic biomarker in colorectal cancer. Gut. 2017;66(1):107–117. doi:10.1136/gutjnl-2015-309359
121. Wei R, Huang GL, Zhang MY, et al. Clinical significance and prognostic value of microRNA expression signatures in hepatocellular carcinoma. Clin Cancer Res. 2013;19(17):4780–4791. doi:10.1158/1078-0432.CCR-12-2728
122. Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi:10.1186/1471-2407-5-64
123. Troester MA, Lee MH, Carter M, et al. Activation of host wound responses in breast cancer microenvironment. Clin Cancer Res. 2009;15(22):7020–7028. doi:10.1158/1078-0432.CCR-09-1126
124. Radovich M, Clare SE, Atale R, et al. Characterizing the heterogeneity of triple-negative breast cancers using microdissected normal ductal epithelium and RNA-sequencing. Breast Cancer Res Treat. 2014;143(1):57–68. doi:10.1007/s10549-013-2780-y
125. Sanz-Pamplona R, Berenguer A, Cordero D, et al. Aberrant gene expression in mucosa adjacent to tumor reveals a molecular crosstalk in colon cancer. Mol Cancer. 2014;13:46. doi:10.1186/1476-4598-13-46
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.