Back to Journals » International Journal of Nanomedicine » Volume 11
The effect of exposure to nanoparticles and nanomaterials on the mammalian epigenome
Authors Sierra MI, Valdés A, Fernández AF , Torrecillas R, Fraga MF
Received 18 August 2016
Accepted for publication 18 October 2016
Published 25 November 2016 Volume 2016:11 Pages 6297—6306
DOI https://doi.org/10.2147/IJN.S120104
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
MI Sierra,1 A Valdés,2 AF Fernández,1 R Torrecillas,2 MF Fraga2
1Cancer Epigenetics Laboratory, Institute of Oncology of Asturias (IUOPA), Hospital Universitario Central de Asturias (HUCA), Universidad de Oviedo, Oviedo, 2Nanomaterials and Nanotechnology Research Center (CINN-CSIC), Universidad de Oviedo-Principado de Asturias, El Entrego, Spain
Abstract: Human exposure to nanomaterials and nanoparticles is increasing rapidly, but their effects on human health are still largely unknown. Epigenetic modifications are attracting ever more interest as possible underlying molecular mechanisms of gene–environment interactions, highlighting them as potential molecular targets following exposure to nanomaterials and nanoparticles. Interestingly, recent research has identified changes in DNA methylation, histone post-translational modifications, and noncoding RNAs in mammalian cells exposed to nanomaterials and nanoparticles. However, the challenge for the future will be to determine the molecular pathways driving these epigenetic alterations, the possible functional consequences, and the potential effects on health.
Keywords: noncoding RNAs, ncRNAs, DNA methylation, histone modification, epigenetics
Introduction
During the last decade, we have witnessed a tremendous increase in the use of nanoparticles and nanomaterials in several areas of human life, chemical industry, food technology, cosmetics and biomedicine, among others (Table 1).1,2 This rise in use is the consequence of developments in nanotechnology, defined according to the US Government’s National Nanotechnology Initiative as “the understanding and control of matter at dimensions of roughly 1 to 100 nm, where unique phenomena enable novel applications.”
Particle sizes up to 100 nm determine changes in the physical and chemical properties of materials compared to those observed at larger scales; for instance, chemical reactivity, electrical conductivity, melting point, fluorescence, and magnetism are all affected by size. This is mainly due to the increase in surface area of nanomaterials compared with larger-scale particles of similar masses, enabling increased contact with the surrounding materials and therefore changes in their reactivity. For example, titanium dioxide (TiO2) particles lose their white color and become colorless at sizes below 50 nm. Other types of particles considered to have electrical insulation properties may become conductive at the nanosize, and substances with low solubility can become more soluble at sizes below 100 nm.3 Furthermore, nanoparticles can combine (homogeneously or heterogeneously) as aggregates or agglomerates; aggregates are formed by particles strongly linked by molecular-type bonds, while agglomerates are formed by particles interlinked by van der Waals-type lower energy bonds.4
Also related to the size of nanoparticles is their coefficient of diffusion, which is inversely proportional to their size and ensures that their main transport mechanism in the environment is air diffusion.4 As a result, independent of their current use in human endeavors, the effects of human exposure to nanoparticles have attracted interest in the last few years from a safety/health point of view. Since it has recently became obvious that the misregulation of cellular epigenetic mechanisms may be detrimental to health, this has further fueled interest in the study of the effects of nanoparticle exposure on epigenetic regulation.5 While epigenetic modifications can be very stable and, in some cases, passed on to multiple generations, they can also change dynamically in response to specific cellular conditions or environmental stimuli. The effect of the environment on the epigenome has been extensively reviewed.6,7 Some authors have already emphasized that the potential epigenetic properties of nanomaterials could not only affect the exposed individuals but also their offspring.8
Here, we will focus on the potential effect of exposure to nanoparticles and nanomaterials on the epigenome, considering epigenetic regulation as the biological mechanism whereby DNA, RNA, and proteins are chemically or structurally modified without changing their primary sequence. These epigenetic modifications play critical roles in the regulation of numerous cellular processes, including gene expression and DNA replication and recombination. Epigenetic regulatory mechanisms include, among others, DNA methylation and hydroxymethylation, histone modification, chromatin remodeling, RNA methylation, and small and long noncoding RNAs (ncRNAs).
There are many different types of nanoparticles and nanomaterials that are available with completely different physical and chemical properties; although there is a natural tendency to assess the physiological effects of nanosubstances in general, in reality, their impact on the organism will be highly dependent on the substance in question. Furthermore, some authors remark that unintentionally emitted nanoparticles in natural systems can be dynamically affected by the environment, complicating even further the assessment of their effect on exposed populations.9
DNA methylation changes in response to exposure to nanoparticles and nanomaterials
DNA methylation consists in the covalent chemical modification of cytosines at the C5 position with a methyl group and occurs at CpG dinucleotides, which are clustered into CpG islands, which affects DNA accessibility to the cellular transcriptional machinery and typically turns off gene expression. This methylation process is catalyzed by DNA methyltransferases (DNMT3a/DNMT3b and DNMT1).10 Demethylation, in contrast, can occur either passively or through active mechanisms in which 10–11 translocator proteins, 5-hydroxymethylcytosine formation, and thymidine glycosidases are all implicated.11
A rapid increase in the use of nanocomposites in the last decade has led several research groups to study alterations in DNA methylation patterns in response to these substances. Examples of nanosubstances studied include carbon-based nanoparticles,12 nano-hydroxyapatite,13 nanoparticles of TiO2 and copper oxides (CuO),14,15 and SiO2.16,17 In general, these studies have identified a number of both locus-specific and global DNA methylation changes in response to nanoparticle and nanomaterial exposure. However, these works provide neither genome-wide data nor correlation with gene expression, which makes it difficult to interpret the extent of DNA methylation changes and their possible functional effects. To our knowledge, the first report on the effect of a nanosized substance on DNA methylation was published only six years ago.16 In this study, the authors incubated the human epidermal keratinocyte cell line HaCaT with increasing concentrations of 15 nm SiO2 for 24 h and then analyzed the global DNA methylation changes using high-performance capillary electrophoresis and immunofluorescence staining. Both assays showed a relative decrease of more than 20% in global DNA methylation. SiO2 was also associated with a decrease in maintenance and de novo DNMTs, leading the authors to propose that deregulation of DNMTs might play an important role in the process. However, such a strong effect in such short period of time could also imply active demethylation processes. Global DNA hypomethylation in response to nanosized substances was also recently observed in vivo.12 In this study, the authors detected significant DNA hypomethylation in the lungs and blood of mice exposed by airways to a type of multiwall carbon nanotubes (MWCN). In contrast to the findings of Gong et al16 with regard to very early DNA methylation changes, the effects were observed only after seven days of exposure in this study.12 Complicating the scenario even further, a recent study using high-performance liquid chromatography-mass spectrometry (HPLC-MS) found an increase in global methylation in a human lung cancer cell line exposed to different concentrations of carbon-based nanoparticles, but a reduction in the expression of DNMTs to different extents depending on the type of carbon nanoparticle.18 Therefore, it is clear that future research is still needed to determine the effect of nanoparticle size and type and the effect of in vivo conditions on the time of exposure required to induce DNA methylation changes.
Global DNA methylation mainly depends on the methylation level of repeated DNA where the majority of the cytosine methylation occurs in the genome, and that is why the level of DNA methylation at certain repeats is widely accepted as a surrogate of global DNA methylation.19 This is of considerable relevance in this context because some recent studies have found a substantial decrease in repeated DNA methylation in response to ambient-air microparticulates exposure.20,21 In contrast, the effect of different types of nanoparticles on the methylome of cultured mammalian cells is less clear. Indeed, a recent study that analyzed the DNA methylation changes at LINE1 and Alu repeats in mammalian cells exposed to different concentrations of printer-emitted engineered nanoparticles and nanoparticles of mild steel welding fumes, CuO, and TiO2 found both hyper- and hypomethylation, depending on the cell type and the repeated DNA sequence involved.14,15 Interestingly, although the DNA methylation changes were small, nanoparticle exposure was clearly associated with DNMT downregulation and repeated DNA reactivation. It is also worth noting that experiments using mouse models have revealed similar methylation changes in response to nanoparticle exposure.22,23 These results highlight the fact that the functional consequences of nanoparticle-dependent DNA methylation changes are still largely unknown and that other molecular mechanisms must be involved in the upregulation of repeated DNA.
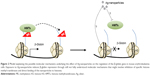
In addition to the methylation changes at the global and repeated DNA level, some studies have identified the changes in response to nanoparticle exposure at specific loci. It should be noted that, generally, promoter hypermethylation is associated with gene silencing, whereas the effect of intragenic methylation is not so clear, although it might also have a role in regulating gene expression.24 A recent work by Ha et al13 reported a 40% increase in DNA methylation at the promoter region of the osteoblast lineage commitment gene alkaline phosphatase (ALPL) in murine bone marrow stromal cells, following treatment with nano-hydroxyapatite, a compound being studied as a therapeutic biomaterial for use as a functional scaffold and implant coating for skeletal repair and dental applications.25,26 An association was found between nano-hydroxyapatite-dependent promoter hypermethylation and ALPL gene repression and, consequently, osteoblast differentiation, which could have important health implications (Figure 1). Although these results are interesting, future research analyzing single CpG methylation at different regions of the ALPL gene is needed to determine the precise role of nanoparticle-dependent DNA methylation changes in gene expression and to determine the molecular mechanism through which nano-hydroxyapatite induces hypermethylation.
In the same vein, promoter-specific hypermethylation of the poly (ADP-ribose) polymerase 1 (PARP-1) was recently reported upon exposure of human lung adenocarcinoma cells to TiO2 nanoparticles.27 PARP-1 is a DNA-binding protein involved in DNA repair, proliferation, and chromatin modification, among other cellular processes. PARP-1 has also been found to form a complex with the DNA methyltransferase DNMT1, the histone H3K9 methyltransferase G9a, and the histone ubiquitin ligase Np95, indicative of a link between polyADP-ribosylation and the epigenome28 and suggesting that it contributes to the genome integrity.29 Treatment with SiO2 nanoparticles has also been found to induce the aberrant hypermethylation and inactivation of PARP-1 in human keratinocytes. Interestingly, DNMT1 knockdown restored PARP-1 promoter methylation levels and its expression,17 further supporting the notion that epigenetic regulations could be involved in the response to SiO2 nanoparticle exposure.
Nanosized substances do not always induce DNA hypermethylation at single-copy sequences. For example, using bisulfite pyrosequencing, Brown et al recently showed a decrease in the promoter methylation levels of the INF-γ and TNF-α genes in the lungs of mice exposed by airways to MWCN.12 In the case of the TNF-α gene, there was a negative correlation between DNA methylation and gene expression, which suggests that DNA methylation might play a role in the regulation of this gene in response to MWCN and, in consequence, in the regulation of the inflammatory response following MWCNT exposure. In another study, researchers identified hypo- and hypermethylation of the differentially methylated regions of the imprinted genes Zac1 and Igf2r, respectively, in mice embryos exposed to silver nanoparticles.30 These alterations might have important implications in disease development, since DNA methylation plays a critical role in the regulation of gene imprinting,31 a critical molecular process during embryonic development. Future research is needed to determine the possible functional consequences of these alterations during embryo development. Silver nanoparticles have also been shown to alter DNA methylation in mouse hippocampus neuronal cells,32 thus suggesting that this type of nanoparticle is an ontogenic-independent modulator of the epigenome.
Therefore, at this moment, it seems clear that, in general, nanoparticle exposure can induce loss of DNA methylation. But, to our knowledge, there are no reports on DNA methylation levels in human populations exposed to specific types of nanoparticles, although there are reports on methylome changes after exposure to ambient-air pollution, where particulate matter (PM) is included at the nano- and micro-scales. A recent study found that exposure to air pollutants (in this case, microparticles of NO2, PM10, PM2.5, and O3) in healthy human individuals led to a small but statistically significant decrease in global DNA methylation.33 However, another study found increased or decreased methylation levels of ten different repeated DNA sequences in men exposed to airborne pollutant.34 The authors attributed this variability in response to the differences in susceptibility of DNMTs to environmental factors, which could modify their methylation activity on specific DNA repeated sequences. Of note is also a recent paper reporting the lack of association between global DNA and ambient PM in dogs.35 The reason for these differences is not clear and, in consequence, further research is needed to establish the factors determining the effect of microparticulates on methylome in vivo. Moreover, considering that most studies have been carried out on peripheral blood and that DNA methylation patterns are in fact cell-type specific, changes in blood cell composition in response to exposure to airborne pollutants could be a confounding factor, and should thus be taken into account in future studies.
Effect of nanomaterials and nanoparticles on histone post-translational modifications
DNA packaging strength is dependent on other epigenetic marks collectively known as histone modifications. There are a variety of such chemical marks (eg, acetylation, methylation, phosphorylation), all of which modify the terminal amino tails of histones, changing how tightly or loosely the DNA is packaged. In general, when the wrapping is tight, a gene is less accessible to the cellular transcription machinery and is consequently expressed less, and when the wrapping is lost, the gene generally becomes more accessible.
The entry of nanoparticles into the cell nucleus can modulate different cellular functions depending on the chromatin region affected; while nanoparticle-mediated heterochromatin changes cause marked shrinkage of the nucleus, euchromatin is only marginally modified.36
The effect of nanosized substances on histone post-translational modifications has been studied much less than their effect on DNA methylation. However, some preliminary studies suggest that histone modifications are also important molecular targets for different types of nanoparticles. For example, a recent study found that the nucleus of human breast cancer cells undergoes chromatin condensation and global histone hypoacetylation after quantum dot treatment with cadmium telluride,37 a nanosubstance currently being considered as a potential diagnostic, therapeutic, and imaging tool. Incubation of MCF-7 cells with cadmium telluride quantum dot for either 4 h or 24 h resulted in global histone H3 hypoacetylation and reduced gene transcription in both cases. Intriguingly, quantum dot treatment increased the expression of some apoptotic genes through the activation of p53. However, the effect of quantum dot exposure on cell death was not investigated in this study. Future research should determine the possible effects of specific nanoparticles on cancer cell death.
Silver nanoparticles have also been recently proposed to affect histone post-translational modifications. These compounds are being utilized in an increasing number of fields and can be found as components of antibacterial coatings, antistatic materials, superconductors, and biosensors. The potential of silver nanoparticles to penetrate inside the nucleus is reported to affect various enzymes involved in chromatin remodeling, such as histone deacetylases.38 In this regard, a recent study showed that sublethal concentrations of silver nanoparticles induced a reduction in hemoglobin levels in mouse erythroleukemia cells, possibly through the diminished methylation of H3K4me3 and H3K79me1.39 Although the underlying molecular mechanisms are not known, the latter authors presented data suggesting that the effect could be mediated by the inhibition of specific histone methyltransferases or even through the binding of silver nanoparticles to histones H3 and H4 (Figure 2).
The shape, or more precisely the topography, of micro- and nanomaterials used as scaffolds for biological and medical applications seems to affect the cellular epigenome, specifically through histone modification alterations. Furthermore, reprogramming somatic cells as pluripotent stem cells is more efficient when bioengineered substrates are used.40 Supporting this notion, a recent study showed that the topography of the cell substrate can modulate cell differentiation and reprogramming.41 Through as yet unknown molecular mechanisms, microgrooved surfaces lead to increased histone H3 acetylation and methylation which is assumed to play an important role in the regulation of cell differentiation and reprogramming processes.41
Effect of exposure to nanoparticles on ncRNA expression
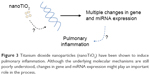
The ncRNAs represent another important component of the epigenetic machinery. They can be classified as short ncRNAs (with less than 200 nucleotides) and long ncRNAs (include all larger transcripts). Furthermore, there are several subtypes of both short (eg, miRNAs, endo-siRNAs, or piRNAs) and long ncRNAs (eg, natural antisense transcripts, long intergenic noncoding RNAs, or sense intronic). Many ncRNAs irrespective of their nature or length are able to modulate gene expression through their interaction with other epigenetic processes, mainly with the expression and function of proteins involved in histone modifications, chromatin remodeling, and/or DNA methylation.42,43 Therefore, many ncRNAs are targets of these same processes.43 It is the plasticity of ncRNAs and their capacity for dynamic interaction with DNA, RNA, and proteins which confers on them the ability to mediate in the various epigenetic processes through which cells respond to diverse external or internal stimuli.42 It is therefore not unexpected that, as with DNA methylation and histone post-translational modification, the biology of ncRNAs can also be affected by exposure to nanosized substances.44 The study by Halappanavar et al was one of the first studies evidencing the effect of nanocompounds on the expression of ncRNAs.45 Using a mouse model, they identified significant changes in the expression of 16 miRNAs in lungs of mice exposed to surface-coated nanotitanium dioxide particles (nanoTiO2), among which mmu-miR-449a presented the greatest change compared with controls. The same study also showed that nanoTiO2 also induced lung inflammation, although the data could not establish a direct link between miRNA alteration and lung injury (Figure 3). Furthermore, a later study from a different group found that pulmonary instillations of Printex® 90, a type of carbon black nanoparticle, resulted in a sustained and substantial induction of three miRNAs in mice for up to one month after exposure. Interestingly, the affected miRNAs identified in each study were different, which raises important questions in relation to the possibility of different nanoparticles targeting specific miRNAs.
In addition to the lungs, nanosized compounds can also affect miRNA expression in other organs in mice. In this regard, a study by Nagano et al identified changes in the blood levels of three liver-specific miRNAs in adult mice exposed to70 nm diameter silica nanoparticles.46 They found that the sensitivity of one of the miRNAs for liver damage was similar to other well-known markers of liver damage. As they can be detected in blood, the authors suggest that altered liver-specific miRNA levels might represent a novel noninvasive biomarker of liver damage in response to nanoparticle exposure. In mice, it has even been proposed that nanoparticle exposure has transplacental epigenetic effects. In their study, Balansky et al found that treatment of pregnant mice with gold nanoparticles (average size of 40–100 nm) induced changes in the expression of 28 miRNAs in fetal tissues.47 As the direct physiological consequences were not assessed, further research is needed to more fully determine the role of nanoparticles in epigenetic regulation during embryonic development, a time period in which epigenetic regulation plays a critical role in processes such us cell fate determination and cell differentiation, among others.48
Till date, the effect of nanoparticle exposure on miRNA expression in humans has only been assessed in vitro. A study by Eom et al showed that exposure of Jurkat T cells to silver nanoparticles altered the expression of 63 miRNAs.49 In line with this effect in vitro, incubation of NIH/3T murine fibroblasts with CdTe quantum dots has been shown to induce significant expression changes in 51 miRNAs.50 In addition, changes in miRNAs, together with chromatin condensation and reorganization, have been reported upon treatment of lung fibroblasts with gold nanoparticles.51 Taken together, the current data suggest that nanomaterial exposure induces alterations in miRNA expression both in vivo and in vitro. However, the underlying molecular mechanisms remain largely unknown.
Conclusion
The rise in the use of nanoparticles in the last decade probably only represents the tip of the iceberg in terms of what the future holds. Given the vast array of fields in which nanoparticles are being used and studied (Table 1), human exposure can only be expected to grow. The epigenetic effects of this exposure are beginning to be studied, but a great deal of work still needs to be done to reveal the whole picture of the impact of such exposure, not only from the epigenetic point of view, where broader epigenomic and mechanistic data are lacking, but also in terms of concrete data on the properties of nanoparticles, composition, size, charge, and functionalization, and whether or not they are degradable or modifiable by external/ambient conditions (Figure 4). Another important challenge for the future will be to determine the possible effect of the genetic variability on the susceptibility to specific nanoparticle exposure.
Acknowledgments
We thank Ronnie Lendrum for editorial assistance. This work has been financially supported by the Plan Nacional de I+D+I 2008–2011/2013–2016/FEDER (PI11/01728 to A.F.F.; PI12/01080 to M.F.F; PI15/00892 to M.F.F.; the ISCIII-Subdirección General de Evaluación y Fomento de la Investigación (Miguel Servet contract: CP11/00131 to A.F.F.); and IUOPA (to M.I.S.). The IUOPA is supported by the Obra Social Cajastur, Spain.
Disclosure
The authors report no conflicts of interest in this work.
References
Leung AW, Kalra J, Santos ND, Bally MB, Anglesio MS. Harnessing the potential of lipid-based nanomedicines for type-specific ovarian cancer treatments. Nanomedicine (Lond). 2014;9(3):501–522. | ||
Loeve S. Of drug administration, war and oïkos: mediating cancer with nanomedicines. Nanomedicine (Lond). 2015;10(21):3261–3274. | ||
Colvin VL. The potential environmental impact of engineered nanomaterials. Nat Biotechnol. 2003;21(10):1166–1170. | ||
Kaluza S, Bladerhaar JK, Orthen B, et al. Work place exposure to nanoparticles. In: Kosk-Bienko J, editor. EU-OSHA Report. European Agency for Safety and Health at Work; 2010. | ||
Martínez E, Lagunas A, Mills CA, et al. Stem cell differentiation by functionalized micro- and nanostructured surfaces. Nanomedicine (Lond). 2009;4(1):65–82. | ||
Feil R, Fraga MF. Epigenetics and the environment: emerging patterns and implications. Nat Rev Genet. 2012;13(2):97–109. | ||
Sierra MI, Fernández AF, Fraga MF. Epigenetics of aging. Curr Genomics. 2015;16:435–440. | ||
Stoccoro A, Karlsson HL, Coppedè F, Migliore L. Epigenetic effects of nano-sized materials. Toxicology. 2013;313(1):3–14. | ||
Yao Y, Costa M. Genetic and epigenetic effects of nanoparticles. J Mol Genet Med. 2013;7:86. | ||
Chen ZX, Riggs AD. DNA methylation and demethylation in mammals. J Biol Chem. 2011;286(21):18347–18353. | ||
Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. | ||
Brown TA, Won Lee J, Holian A, et al. Alterations in DNA methylation corresponding with lung inflammation and as a biomarker for disease development after MWCNT exposure. Nanotoxicology. 2016;10(4):453–461. | ||
Ha SW, Jang HL, Nam KT, Beck GR Jr. Nano-hydroxyapatite modulates osteoblast lineage commitment by stimulation of DNA methylation and regulation of gene expression. Biomaterials. 2015;65:32–42. | ||
Lu X, Miousse IR, Pirela SV, Melnyk S, Koturbash I, Demokritou P. Short-term exposure to engineered nanomaterials affects cellular epigenome. Nanotoxicology. 2016;10(2):140–150. | ||
Pirela SP, Miousse IR, Lu X, et al. Effects of laser printer–emitted engineered nanoparticles on cytotoxicity, chemokine expression, reactive oxygen species, DNA methylation, and DNA damage: a comprehensive in vitro analysis in human small airway epithelial cells, macrophages, and lymphoblasts. Environ Health Perspect. 2016;124(2):210–219. | ||
Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. SiO(2) nanoparticles induce global genomic hypomethylation in HaCaT cells. Biochem Biophys Res Commun. 2010;397(3):397–400. | ||
Gong C, Tao G, Yang L, Liu J, Liu Q, Zhuang Z. Methylation of PARP-1 promoter involved in the regulation of nano-SiO2-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2012;209(3):264–269. | ||
Li J, Tian M, Cui L, et al. Low-dose carbon-based nanoparticle-induced effects in A549 lung cells determined by biospectroscopy are associated with increases in genomic methylation. Sci Rep. 2016;6:20207. | ||
Yang AS, Estécio MRH, Doshi K, Kondo Y, Tajara EH, Issa JPJ. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. | ||
Baccarelli A, Tarantini L, Wright RO, et al. Repetitive element DNA methylation and circulating endothelial and inflammation markers in the VA Normative Aging Study. Epigenetics. 2010;5(3):222–228. | ||
Madrigano J, Baccarelli A, Mittleman MA, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways and DNA methylation in a cohort of older men. Environ Health Perspect. 2011;119(7):977–982. | ||
Lu X, Miousse IR, Pirela SV, et al. In vivo epigenetic effects induced by engineered nanomaterials: a case study of copper oxide and laser printer-emitted engineered nanoparticles. Nanotoxicology. 2016;10(5):629–639. | ||
Pirela SP, Lu X, Miousse IR, et al. Effects of intratracheally instilled lase printer-emitted engineered nanoparticles in a mouse model: a case study of toxicological implications from nanomaterials released during consumer use. NanoImpact. 2016;1:1–8. | ||
Ball MP, Li JB, Gao Y, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27(4):361–368. | ||
Zhou H, Lee J. Nanoscale hydroxyapatite particles for bone tissue engineering. Acta Biomater. 2011;7(7):2769–2781. | ||
Zhang JZ, Liu GY, Wu QO, Zuo JL, Qin YG, Wang JC. Novel mesoporous hydroxyapatite/chitosan composite for bone repair. J Bionic Eng. 2012;9(2):243–251. | ||
Bai W, Chen Y, Gao A. Cross talk between poly(ADP-ribose) polymerase 1 methylation and oxidative stress involved in the toxic effect of anatase titanium dioxide nanoparticles. Int J Nanomed. 2015;10:5561–5569. | ||
Sharif J, Muto M, Takebayashi S, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450(7171):908–912. | ||
Veith S, Mangerich A. RecQ helicases and PARP1 team up in maintaining genome integrity. Ageing Res Rev. 2015;23(Pt A):12–28. | ||
Zhang XF, Park JH, Choi YJ, Kang MH, Gurunathan S, Kim JH. Silver nanoparticles cause complications in pregnant mice. Int J Nanomed. 2015;10:7057–7071. | ||
Voon HP, Gibbons RJ. Maintaining memory of silencing at imprinted differentially methylated regions. Cell Mol Life Sci. 2016;79(3):1871–1879. | ||
Mytych J, Zebrosky J, Lewinska A, Wnuk M. Prolonged effects of silver nanoparticles on p53/p21 pathway-mediated proliferation, DNA damage response, and methylation parameters in HT22 hippocampal neuronal cells. Mol Neurobiol. Epub 2016 Feb 3. | ||
de Prins S, Koppen G, Jacobs G, et al. Influence of ambient air pollution on global DNA methylation in healthy adults: a seasonal follow-up. Environ Int. 2013;59:418–424. | ||
Byun H-M, Motta V, Panni T, et al. Evolutionary age of repetitive element subfamilies and sensitivity of DNA methylation to airborne pollutants. Part Fibre Toxicol. 2013;10:28–37. | ||
Montrose L, Noonan C, Cho YH, et al. Evaluating the effect of ambient particulate pollution on DNA methylation in Alaskan sled dogs: potential applications for a sentinel model of human health. Sci Total Environ. 2015;512–513:489–494. | ||
Mytych J, Wnuk M. Nanoparticle technology as a double-edged sword: cytotoxic, genotoxic and epigenetic effects on living cells. J Biomater Nanobiotechnol. 2013;4:53–63. | ||
Choi AO, Brown SE, Szyf M, Maysinger D. Quantum dot-induced epigenetic and genotoxic changes in human breast cancer cells. J Mol Med (Berl). 2008;86(3):291–302. | ||
Dubey P, Matai I, Kumar SU, Sachdev A, Bhushana B, Gopinath P. Perturbation of cellular mechanistic system by silver nanoparticle toxicity: cytotoxic, genotoxic and epigenetic potentials. Adv Colloid Interface Sci. 2015;221:4–21. | ||
Qian Y, Zhang J, Hu Q, et al. Silver nanoparticle-induced hemoglobin decrease involves alteration of histone 3 methylation status. Biomaterials. 2015;70:12–22. | ||
Tong Z, Solanki A, Hamilos A, et al. Application of biomaterials to advance induced pluripotent stem cell research and therapy. EMBO J. 2015;34(8):987–1008. | ||
Downing TL, Soto J, Morez C, et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12(12):1154–1162. | ||
Zhao Y, Sun H, Wang H. Long noncoding RNAs in DNA methylation: new players stepping into the old game. Cell Biosci. 2016;6:45. | ||
Peschansky VJ, Wahlestedt C. Non-coding RNAs as direct and indirect modulators of epigenetic regulation. Epigenetics. 2014;9(1):3–12. | ||
Vrijens K, Bollati V, Nawrot TS. MicroRNAs as potential signatures of environ-mental exposure or effect: a systematic review. Environ Health Perspect. 2015;123(5):399–411. | ||
Halappanavar S, Jackson P, Williams A, et al. Pulmonary response to surface-coated nanotitanium dioxide particles includes induction of acute phase response genes, inflammatory cascades, and changes in microRNAs: a toxicogenomic study. Environ Mol Mutagen. 2011;52(6):425–439. | ||
Nagano T, Higashisaka K, Kunieda A, et al. Liver-specific microRNAs as biomarkers of nanomaterial-induced liver damage. Nanotechnology. 2013;24(40):405102. | ||
Balansky R, Longobardi M, Ganchev G, et al. Transplacental clastogenic and epigenetic effects of gold nanoparticles in mice. Mutat Res. 2013;751–752:42–48. | ||
Hochberg Z, Feil R, Constancia M, et al. Child health, developmental plasticity, and epigenetic programming. Endocr Rev. 2011;32(2):159–224. | ||
Eom HJ, Chatterjee N, Lee J, Choi J. Integrated mRNA and micro RNA profiling reveals epigenetic mechnism of differential sensitivity of Jurkat T cells to AGNPs and Ag ions. Toxicol Lett. 2014;229(1):311–318. | ||
Li S, Wang H, Qi Y, et al. Assessment of nanomaterial cytotoxicity with SOLiD sequencing-based microRNA expression profiling. Biomaterials. 2011;32(34):9021–9030. | ||
Ng CT, Dheen ST, Yip WCG, Ong CN, Bay BH, Lanry Yung LY. The induction of epigenetic regulation of PROS1 gene in lung fibroblasts by gold nanoparticles and implications for potential lung injury. Biomaterials. 2011;32(30):7609–7615. | ||
Braydich-Stolle LK, Lucas B, Schrand A, et al. Silver nanoparticles disrupt GDN/Fyn kinase signaling in spermatogonial stem cells. Toxicol Sci. 2010;116(2):577–589. | ||
Conroy J, Byrne SJ, Gun’ko YK, et al. CdTe nanoparticles display tropism to core histones and histone-rich cell organelles. Small. 2008;4(11):2006–2015. | ||
Tucci P, Porta G, Agostini M, et al. Metabolic effects of TiO2 nanoparticles, a common component of sunscreens and cosmetics, on human keratinocytes. Cell Death Dis. 2013;4:e549. | ||
Patil NA, Gade WN, Deobagkar SS. Epigenetic modulation upon exposure of lung fibroblasts to TiO2 and ZnO nanoparticles: alterations in DNA methylation. Int J Nanomed. 2016;11:4509–4519. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.